| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Lysophosphatidylserine lipase ABHD12 |
|---|
| Ligand | BDBM50556537 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2056359 (CHEMBL4711360) |
|---|
| IC50 | 600±n/a nM |
|---|
| Citation |  Arena, C; Gado, F; Di Cesare Mannelli, L; Cervetto, C; Carpi, S; Reynoso-Moreno, I; Polini, B; Vallini, E; Chicca, S; Lucarini, E; Bertini, S; D'Andrea, F; Digiacomo, M; Poli, G; Tuccinardi, T; Macchia, M; Gertsch, J; Marcoli, M; Nieri, P; Ghelardini, C; Chicca, A; Manera, C The endocannabinoid system dual-target ligand N-cycloheptyl-1,2-dihydro-5-bromo-1-(4-fluorobenzyl)-6-methyl-2-oxo-pyridine-3-carboxamide improves disease severity in a mouse model of multiple sclerosis. Eur J Med Chem208:0 (2020) [PubMed] Article Arena, C; Gado, F; Di Cesare Mannelli, L; Cervetto, C; Carpi, S; Reynoso-Moreno, I; Polini, B; Vallini, E; Chicca, S; Lucarini, E; Bertini, S; D'Andrea, F; Digiacomo, M; Poli, G; Tuccinardi, T; Macchia, M; Gertsch, J; Marcoli, M; Nieri, P; Ghelardini, C; Chicca, A; Manera, C The endocannabinoid system dual-target ligand N-cycloheptyl-1,2-dihydro-5-bromo-1-(4-fluorobenzyl)-6-methyl-2-oxo-pyridine-3-carboxamide improves disease severity in a mouse model of multiple sclerosis. Eur J Med Chem208:0 (2020) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Lysophosphatidylserine lipase ABHD12 |
|---|
| Name: | Lysophosphatidylserine lipase ABHD12 |
|---|
| Synonyms: | ABD12_HUMAN | ABHD12 | C20orf22 | Monoacylglycerol lipase ABHD12 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 45111.32 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_1514755 |
|---|
| Residue: | 398 |
|---|
| Sequence: | MRKRTEPVALEHERCAAAGSSSSGSAAAALDADCRLKQNLRLTGPAAAEPRCAADAGMKR
ALGRRKGVWLRLRKILFCVLGLYIAIPFLIKLCPGIQAKLIFLNFVRVPYFIDLKKPQDQ
GLNHTCNYYLQPEEDVTIGVWHTVPAVWWKNAQGKDQMWYEDALASSHPIILYLHGNAGT
RGGDHRVELYKVLSSLGYHVVTFDYRGWGDSVGTPSERGMTYDALHVFDWIKARSGDNPV
YIWGHSLGTGVATNLVRRLCERETPPDALILESPFTNIREEAKSHPFSVIYRYFPGFDWF
FLDPITSSGIKFANDENVKHISCPLLILHAEDDPVVPFQLGRKLYSIAAPARSFRDFKVQ
FVPFHSDLGYRHKYIYKSPELPRILREFLGKSEPEHQH
|
|
|
|---|
| BDBM50556537 |
|---|
| n/a |
|---|
| Name | BDBM50556537 |
|---|
| Synonyms: | CHEMBL4757488 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H28F2N2O2 |
|---|
| Mol. Mass. | 450.5202 |
|---|
| SMILES | Cc1c(cc(C(=O)NC2CCCCCC2)c(=O)n1Cc1ccc(F)cc1)-c1ccc(F)cc1 |
|---|
| Structure | 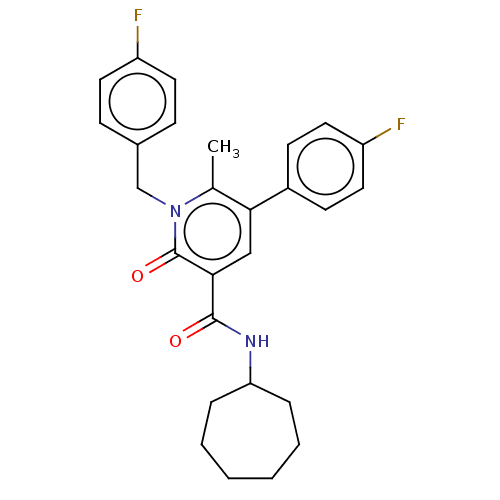
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Arena, C; Gado, F; Di Cesare Mannelli, L; Cervetto, C; Carpi, S; Reynoso-Moreno, I; Polini, B; Vallini, E; Chicca, S; Lucarini, E; Bertini, S; D'Andrea, F; Digiacomo, M; Poli, G; Tuccinardi, T; Macchia, M; Gertsch, J; Marcoli, M; Nieri, P; Ghelardini, C; Chicca, A; Manera, C The endocannabinoid system dual-target ligand N-cycloheptyl-1,2-dihydro-5-bromo-1-(4-fluorobenzyl)-6-methyl-2-oxo-pyridine-3-carboxamide improves disease severity in a mouse model of multiple sclerosis. Eur J Med Chem208:0 (2020) [PubMed] Article
Arena, C; Gado, F; Di Cesare Mannelli, L; Cervetto, C; Carpi, S; Reynoso-Moreno, I; Polini, B; Vallini, E; Chicca, S; Lucarini, E; Bertini, S; D'Andrea, F; Digiacomo, M; Poli, G; Tuccinardi, T; Macchia, M; Gertsch, J; Marcoli, M; Nieri, P; Ghelardini, C; Chicca, A; Manera, C The endocannabinoid system dual-target ligand N-cycloheptyl-1,2-dihydro-5-bromo-1-(4-fluorobenzyl)-6-methyl-2-oxo-pyridine-3-carboxamide improves disease severity in a mouse model of multiple sclerosis. Eur J Med Chem208:0 (2020) [PubMed] Article