

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cytochrome P450 3A4 | ||
| Ligand | BDBM50598849 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_2228092 (CHEMBL5141605) | ||
| IC50 | 23000±n/a nM | ||
| Citation |  Szychowski, J; Papp, R; Dietrich, E; Liu, B; Vallée, F; Leclaire, ME; Fourtounis, J; Martino, G; Perryman, AL; Pau, V; Yin, SY; Mader, P; Roulston, A; Truchon, JF; Marshall, CG; Diallo, M; Duffy, NM; Stocco, R; Godbout, C; Bonneau-Fortin, A; Kryczka, R; Bhaskaran, V; Mao, D; Orlicky, S; Beaulieu, P; Turcotte, P; Kurinov, I; Sicheri, F; Mamane, Y; Gallant, M; Black, WC Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306. J Med Chem65:10251-10284 (2022) [PubMed] Article Szychowski, J; Papp, R; Dietrich, E; Liu, B; Vallée, F; Leclaire, ME; Fourtounis, J; Martino, G; Perryman, AL; Pau, V; Yin, SY; Mader, P; Roulston, A; Truchon, JF; Marshall, CG; Diallo, M; Duffy, NM; Stocco, R; Godbout, C; Bonneau-Fortin, A; Kryczka, R; Bhaskaran, V; Mao, D; Orlicky, S; Beaulieu, P; Turcotte, P; Kurinov, I; Sicheri, F; Mamane, Y; Gallant, M; Black, WC Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306. J Med Chem65:10251-10284 (2022) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cytochrome P450 3A4 | |||
| Name: | Cytochrome P450 3A4 | ||
| Synonyms: | Albendazole monooxygenase | Albendazole sulfoxidase | CP3A4_HUMAN | CYP3A3 | CYP3A4 | CYPIIIA3 | CYPIIIA4 | Cytochrome P450 3A3 | Cytochrome P450 3A4 (CYP3A4) | Cytochrome P450 HLp | Nifedipine oxidase | Quinine 3-monooxygenase | Taurochenodeoxycholate 6-alpha-hydroxylase | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 57349.57 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 503 | ||
| Sequence: |
| ||
| BDBM50598849 | |||
| n/a | |||
| Name | BDBM50598849 | ||
| Synonyms: | CHEMBL5204520 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C18H16N6O2S | ||
| Mol. Mass. | 380.424 | ||
| SMILES | Cc1ccc(O)c(C)c1-n1c(N)c(C(N)=O)c2nc(cnc12)-c1nccs1 |(3.88,-2.75,;4.17,-1.55,;5.65,-1.12,;6.01,.38,;4.9,1.44,;5.19,2.64,;3.42,1.01,;2.53,1.86,;3.06,-.49,;1.55,-.81,;.93,-2.21,;1.54,-3.28,;-.61,-2.05,;-1.63,-3.2,;-2.84,-2.94,;-1.25,-4.37,;-.92,-.55,;-2.26,.22,;-2.26,1.76,;-.92,2.53,;.41,1.76,;.41,.22,;-3.59,2.53,;-3.74,4.05,;-5.24,4.37,;-6.01,3.04,;-4.98,1.89,)| | ||
| Structure | 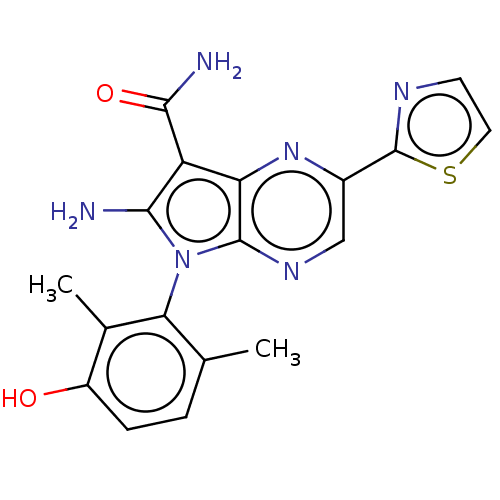
| ||