

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Bile acid receptor | ||
| Ligand | BDBM50572221 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_2234712 (CHEMBL5148484) | ||
| EC50 | 320±n/a nM | ||
| Citation |  Nara, SJ; Jogi, S; Cheruku, S; Kandhasamy, S; Jaipuri, F; Kathi, PK; Reddy, S; Sarodaya, S; Cook, EM; Wang, T; Sitkoff, D; Rossi, KA; Ruzanov, M; Kiefer, SE; Khan, JA; Gao, M; Reddy, S; Sivaprasad Lvj, S; Sane, R; Mosure, K; Zhuo, X; Cao, GG; Ziegler, M; Azzara, A; Krupinski, J; Soars, MG; Ellsworth, BA; Wacker, DA Discovery of BMS-986339, a Pharmacologically Differentiated Farnesoid X Receptor Agonist for the Treatment of Nonalcoholic Steatohepatitis. J Med Chem65:8948-8960 (2022) [PubMed] Article Nara, SJ; Jogi, S; Cheruku, S; Kandhasamy, S; Jaipuri, F; Kathi, PK; Reddy, S; Sarodaya, S; Cook, EM; Wang, T; Sitkoff, D; Rossi, KA; Ruzanov, M; Kiefer, SE; Khan, JA; Gao, M; Reddy, S; Sivaprasad Lvj, S; Sane, R; Mosure, K; Zhuo, X; Cao, GG; Ziegler, M; Azzara, A; Krupinski, J; Soars, MG; Ellsworth, BA; Wacker, DA Discovery of BMS-986339, a Pharmacologically Differentiated Farnesoid X Receptor Agonist for the Treatment of Nonalcoholic Steatohepatitis. J Med Chem65:8948-8960 (2022) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Bile acid receptor | |||
| Name: | Bile acid receptor | ||
| Synonyms: | Bar | Farnesoid X-activated receptor | Farnesol receptor HRR-1 | Fxr | NR1H4_MOUSE | Nr1h4 | Nuclear receptor subfamily 1 group H member 4 | RXR-interacting protein 14 | Retinoid X receptor-interacting protein 14 | Rip14 | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 55996.68 | ||
| Organism: | Mus musculus | ||
| Description: | ChEMBL_793021 | ||
| Residue: | 488 | ||
| Sequence: |
| ||
| BDBM50572221 | |||
| n/a | |||
| Name | BDBM50572221 | ||
| Synonyms: | CHEMBL4862974 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C30H23Cl2F3N4O3 | ||
| Mol. Mass. | 615.43 | ||
| SMILES | OC(=O)c1cc(c2cc(ccc2n1)N1CCC2(CC(=C2)c2c(onc2-c2c(Cl)cncc2Cl)C2CC2)CC1)C(F)(F)F |c:20,(46.96,-23.25,;46.47,-24.71,;47.49,-25.87,;44.96,-25.01,;44.46,-26.48,;42.95,-26.78,;41.94,-25.62,;40.43,-25.91,;39.43,-24.74,;39.93,-23.29,;41.43,-22.99,;42.44,-24.16,;43.95,-23.86,;37.92,-25.03,;37.41,-26.48,;35.91,-26.76,;34.9,-25.6,;33.89,-24.44,;32.72,-25.45,;33.74,-26.62,;31.19,-25.35,;30.71,-23.88,;29.16,-23.89,;28.69,-25.36,;29.95,-26.26,;29.96,-27.8,;28.63,-28.57,;27.29,-27.8,;28.63,-30.11,;29.96,-30.88,;31.3,-30.11,;31.29,-28.56,;32.62,-27.79,;31.6,-22.64,;33.01,-22.01,;31.76,-21.1,;35.4,-24.15,;36.91,-23.86,;42.45,-28.23,;43.47,-29.39,;40.97,-27.83,;41.68,-29.57,)| | ||
| Structure | 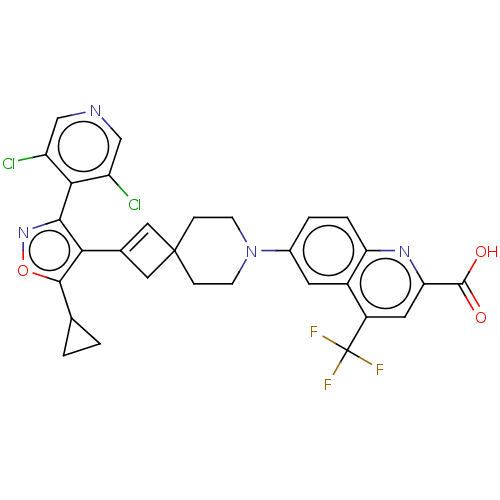
| ||