| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Mitogen-activated protein kinase 7 |
|---|
| Ligand | BDBM50605524 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2251875 (CHEMBL5166085) |
|---|
| IC50 | 25±n/a nM |
|---|
| Citation |  Miller, DC; Reuillon, T; Molyneux, L; Blackburn, T; Cook, SJ; Edwards, N; Endicott, JA; Golding, BT; Griffin, RJ; Hardcastle, I; Harnor, SJ; Heptinstall, A; Lochhead, P; Martin, MP; Martin, NC; Myers, S; Newell, DR; Noble, RA; Phillips, N; Rigoreau, L; Thomas, H; Tucker, JA; Wang, LZ; Waring, MJ; Wong, AC; Wedge, SR; Noble, MEM; Cano, C Parallel Optimization of Potency and Pharmacokinetics Leading to the Discovery of a Pyrrole Carboxamide ERK5 Kinase Domain Inhibitor. J Med Chem65:6513-6540 (2022) [PubMed] Article Miller, DC; Reuillon, T; Molyneux, L; Blackburn, T; Cook, SJ; Edwards, N; Endicott, JA; Golding, BT; Griffin, RJ; Hardcastle, I; Harnor, SJ; Heptinstall, A; Lochhead, P; Martin, MP; Martin, NC; Myers, S; Newell, DR; Noble, RA; Phillips, N; Rigoreau, L; Thomas, H; Tucker, JA; Wang, LZ; Waring, MJ; Wong, AC; Wedge, SR; Noble, MEM; Cano, C Parallel Optimization of Potency and Pharmacokinetics Leading to the Discovery of a Pyrrole Carboxamide ERK5 Kinase Domain Inhibitor. J Med Chem65:6513-6540 (2022) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Mitogen-activated protein kinase 7 |
|---|
| Name: | Mitogen-activated protein kinase 7 |
|---|
| Synonyms: | BMK1 | Big MAP kinase 1 | ERK5 | Extracellular signal-regulated kinase 5 (ERK5) | MAPK7 | MK07_HUMAN | PRKM7 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 88377.36 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q13164 |
|---|
| Residue: | 816 |
|---|
| Sequence: | MAEPLKEEDGEDGSAEPPGPVKAEPAHTAASVAAKNLALLKARSFDVTFDVGDEYEIIET
IGNGAYGVVSSARRRLTGQQVAIKKIPNAFDVVTNAKRTLRELKILKHFKHDNIIAIKDI
LRPTVPYGEFKSVYVVLDLMESDLHQIIHSSQPLTLEHVRYFLYQLLRGLKYMHSAQVIH
RDLKPSNLLVNENCELKIGDFGMARGLCTSPAEHQYFMTEYVATRWYRAPELMLSLHEYT
QAIDLWSVGCIFGEMLARRQLFPGKNYVHQLQLIMMVLGTPSPAVIQAVGAERVRAYIQS
LPPRQPVPWETVYPGADRQALSLLGRMLRFEPSARISAAAALRHPFLAKYHDPDDEPDCA
PPFDFAFDREALTRERIKEAIVAEIEDFHARREGIRQQIRFQPSLQPVASEPGCPDVEMP
SPWAPSGDCAMESPPPAPPPCPGPAPDTIDLTLQPPPPVSEPAPPKKDGAISDNTKAALK
AALLKSLRSRLRDGPSAPLEAPEPRKPVTAQERQREREEKRRRRQERAKEREKRRQERER
KERGAGASGGPSTDPLAGLVLSDNDRSLLERWTRMARPAAPALTSVPAPAPAPTPTPTPV
QPTSPPPGPVAQPTGPQPQSAGSTSGPVPQPACPPPGPAPHPTGPPGPIPVPAPPQIATS
TSLLAAQSLVPPPGLPGSSTPGVLPYFPPGLPPPDAGGAPQSSMSESPDVNLVTQQLSKS
QVEDPLPPVFSGTPKGSGAGYGVGFDLEEFLNQSFDMGVADGPQDGQADSASLSASLLAD
WLEGHGMNPADIESLQREIQMDSPMLLADLPDLQDP
|
|
|
|---|
| BDBM50605524 |
|---|
| n/a |
|---|
| Name | BDBM50605524 |
|---|
| Synonyms: | CHEMBL5193573 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H24Cl2FN5O2 |
|---|
| Mol. Mass. | 504.384 |
|---|
| SMILES | CN(C1CCN(C)CC1)c1ccc(NC(=O)c2cc(c[nH]2)C(=O)c2c(Cl)ccc(Cl)c2F)cn1 |
|---|
| Structure | 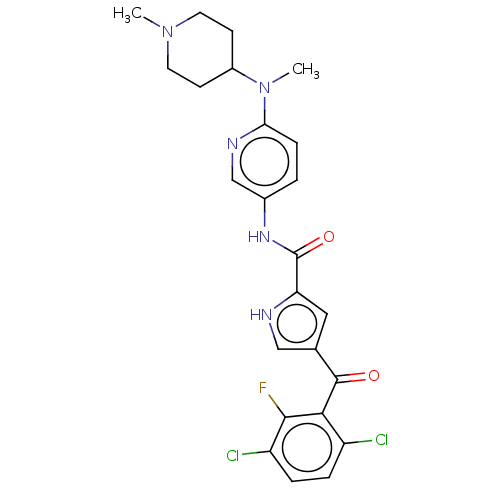
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Miller, DC; Reuillon, T; Molyneux, L; Blackburn, T; Cook, SJ; Edwards, N; Endicott, JA; Golding, BT; Griffin, RJ; Hardcastle, I; Harnor, SJ; Heptinstall, A; Lochhead, P; Martin, MP; Martin, NC; Myers, S; Newell, DR; Noble, RA; Phillips, N; Rigoreau, L; Thomas, H; Tucker, JA; Wang, LZ; Waring, MJ; Wong, AC; Wedge, SR; Noble, MEM; Cano, C Parallel Optimization of Potency and Pharmacokinetics Leading to the Discovery of a Pyrrole Carboxamide ERK5 Kinase Domain Inhibitor. J Med Chem65:6513-6540 (2022) [PubMed] Article
Miller, DC; Reuillon, T; Molyneux, L; Blackburn, T; Cook, SJ; Edwards, N; Endicott, JA; Golding, BT; Griffin, RJ; Hardcastle, I; Harnor, SJ; Heptinstall, A; Lochhead, P; Martin, MP; Martin, NC; Myers, S; Newell, DR; Noble, RA; Phillips, N; Rigoreau, L; Thomas, H; Tucker, JA; Wang, LZ; Waring, MJ; Wong, AC; Wedge, SR; Noble, MEM; Cano, C Parallel Optimization of Potency and Pharmacokinetics Leading to the Discovery of a Pyrrole Carboxamide ERK5 Kinase Domain Inhibitor. J Med Chem65:6513-6540 (2022) [PubMed] Article