

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Sodium/glucose cotransporter 2 | ||
| Ligand | BDBM50265219 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_496716 (CHEMBL1005353) | ||
| IC50 | 835±n/a nM | ||
| Citation |  Lansdell, MI; Burring, DJ; Hepworth, D; Strawbridge, M; Graham, E; Guyot, T; Betson, MS; Hart, JD Design and synthesis of fluorescent SGLT2 inhibitors. Bioorg Med Chem Lett18:4944-7 (2008) [PubMed] Article Lansdell, MI; Burring, DJ; Hepworth, D; Strawbridge, M; Graham, E; Guyot, T; Betson, MS; Hart, JD Design and synthesis of fluorescent SGLT2 inhibitors. Bioorg Med Chem Lett18:4944-7 (2008) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Sodium/glucose cotransporter 2 | |||
| Name: | Sodium/glucose cotransporter 2 | ||
| Synonyms: | Na(+)/glucose cotransporter 2 | SC5A2_HUMAN | SGLT2 | SLC5A2 | Sodium-Dependent Glucose Cotransporter 2 (SGLT2) | Sodium/glucose cotransporter 1 (SGLT1) | Solute carrier family 5 member 2 | ||
| Type: | Protein | ||
| Mol. Mass.: | 72902.00 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P31639 | ||
| Residue: | 672 | ||
| Sequence: |
| ||
| BDBM50265219 | |||
| n/a | |||
| Name | BDBM50265219 | ||
| Synonyms: | 5-(5-(4-(2-chloro-5-((2S,3R,4R,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)benzyl)phenoxy)pentanoyl)-2-(6-hydroxy-3-oxo-3H-xanthen-9-yl)benzoic acid | CHEMBL449778 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C44H39ClO12 | ||
| Mol. Mass. | 795.226 | ||
| SMILES | OC[C@H]1O[C@H]([C@H](O)[C@@H](O)[C@@H]1O)c1ccc(Cl)c(Cc2ccc(OCCCCC(=O)c3ccc(c(c3)C(O)=O)-c3c4ccc(O)cc4oc4cc(=O)ccc34)cc2)c1 |r,wU:2.1,7.7,4.11,wD:5.5,9.10,(-7.73,-4.49,;-6.4,-3.72,;-5.06,-4.48,;-3.73,-3.71,;-2.4,-4.48,;-2.4,-6.03,;-1.07,-6.8,;-3.73,-6.79,;-3.73,-8.33,;-5.06,-6.03,;-6.39,-6.8,;-1.06,-3.72,;-1.06,-2.18,;.28,-1.4,;1.61,-2.18,;2.95,-1.41,;1.61,-3.72,;2.95,-4.49,;4.29,-3.73,;5.6,-4.51,;6.93,-3.76,;6.94,-2.24,;8.28,-1.48,;9.61,-2.26,;10.95,-1.5,;12.28,-2.28,;13.62,-1.52,;14.95,-2.31,;14.97,-3.85,;16.28,-1.53,;16.26,.01,;17.59,.79,;18.93,.03,;18.94,-1.52,;17.61,-2.29,;19.7,-2.86,;18.91,-4.18,;21.24,-2.87,;20.26,.81,;20.25,2.35,;18.92,3.1,;18.9,4.63,;20.22,5.41,;20.21,6.95,;21.55,4.65,;21.56,3.13,;22.91,2.38,;22.92,.83,;24.26,.08,;24.28,-1.46,;25.62,-2.22,;22.95,-2.26,;21.61,-1.5,;21.59,.05,;5.62,-1.46,;4.3,-2.22,;.27,-4.48,)| | ||
| Structure | 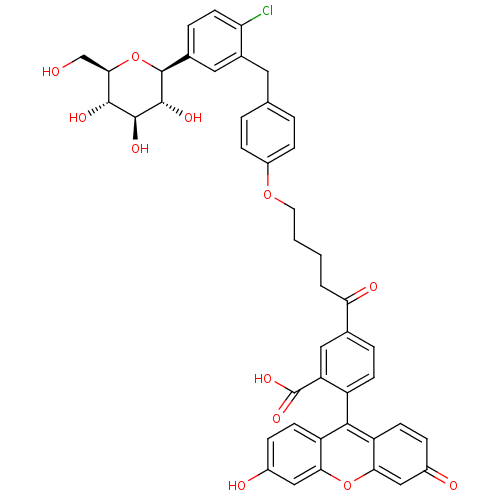
| ||