| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Metabotropic glutamate receptor 2 |
|---|
| Ligand | BDBM50277805 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_502279 (CHEMBL983950) |
|---|
| EC50 | 15±n/a nM |
|---|
| Citation |  Duplantier, AJ; Efremov, I; Candler, J; Doran, AC; Ganong, AH; Haas, JA; Hanks, AN; Kraus, KG; Lazzaro, JT; Lu, J; Maklad, N; McCarthy, SA; O'Sullivan, TJ; Rogers, BN; Siuciak, JA; Spracklin, DK; Zhang, L 3-Benzyl-1,3-oxazolidin-2-ones as mGluR2 positive allosteric modulators: Hit-to lead and lead optimization. Bioorg Med Chem Lett19:2524-9 (2009) [PubMed] Article Duplantier, AJ; Efremov, I; Candler, J; Doran, AC; Ganong, AH; Haas, JA; Hanks, AN; Kraus, KG; Lazzaro, JT; Lu, J; Maklad, N; McCarthy, SA; O'Sullivan, TJ; Rogers, BN; Siuciak, JA; Spracklin, DK; Zhang, L 3-Benzyl-1,3-oxazolidin-2-ones as mGluR2 positive allosteric modulators: Hit-to lead and lead optimization. Bioorg Med Chem Lett19:2524-9 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Metabotropic glutamate receptor 2 |
|---|
| Name: | Metabotropic glutamate receptor 2 |
|---|
| Synonyms: | GPRC1B | GRM2 | GRM2_HUMAN | MGLUR2 | Metabotropic glutamate receptor | glutamate receptor, metabotropic 2 precursor | mGlu2 | metabotropic glutamate 2 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 95584.88 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q14416 |
|---|
| Residue: | 872 |
|---|
| Sequence: | MGSLLALLALLLLWGAVAEGPAKKVLTLEGDLVLGGLFPVHQKGGPAEDCGPVNEHRGIQ
RLEAMLFALDRINRDPHLLPGVRLGAHILDSCSKDTHALEQALDFVRASLSRGADGSRHI
CPDGSYATHGDAPTAITGVIGGSYSDVSIQVANLLRLFQIPQISYASTSAKLSDKSRYDY
FARTVPPDFFQAKAMAEILRFFNWTYVSTVASEGDYGETGIEAFELEARARNICVATSEK
VGRAMSRAAFEGVVRALLQKPSARVAVLFTRSEDARELLAASQRLNASFTWVASDGWGAL
ESVVAGSEGAAEGAITIELASYPISDFASYFQSLDPWNNSRNPWFREFWEQRFRCSFRQR
DCAAHSLRAVPFEQESKIMFVVNAVYAMAHALHNMHRALCPNTTRLCDAMRPVNGRRLYK
DFVLNVKFDAPFRPADTHNEVRFDRFGDGIGRYNIFTYLRAGSGRYRYQKVGYWAEGLTL
DTSLIPWASPSAGPLPASRCSEPCLQNEVKSVQPGEVCCWLCIPCQPYEYRLDEFTCADC
GLGYWPNASLTGCFELPQEYIRWGDAWAVGPVTIACLGALATLFVLGVFVRHNATPVVKA
SGRELCYILLGGVFLCYCMTFIFIAKPSTAVCTLRRLGLGTAFSVCYSALLTKTNRIARI
FGGAREGAQRPRFISPASQVAICLALISGQLLIVVAWLVVEAPGTGKETAPERREVVTLR
CNHRDASMLGSLAYNVLLIALCTLYAFKTRKCPENFNEAKFIGFTMYTTCIIWLAFLPIF
YVTSSDYRVQTTTMCVSVSLSGSVVLGCLFAPKLHIILFQPQKNVVSHRAPTSRFGSAAA
RASSSLGQGSGSQFVPTVCNGREVVDSTTSSL
|
|
|
|---|
| BDBM50277805 |
|---|
| n/a |
|---|
| Name | BDBM50277805 |
|---|
| Synonyms: | 3-(4-(cyclohexylmethoxy)benzyl)-5-methyloxazolidin-2-one | CHEMBL450207 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H25NO3 |
|---|
| Mol. Mass. | 303.396 |
|---|
| SMILES | CC1CN(Cc2ccc(OCC3CCCCC3)cc2)C(=O)O1 |
|---|
| Structure | 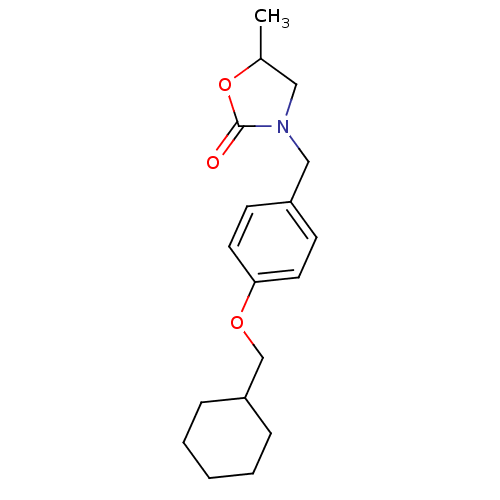
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Duplantier, AJ; Efremov, I; Candler, J; Doran, AC; Ganong, AH; Haas, JA; Hanks, AN; Kraus, KG; Lazzaro, JT; Lu, J; Maklad, N; McCarthy, SA; O'Sullivan, TJ; Rogers, BN; Siuciak, JA; Spracklin, DK; Zhang, L 3-Benzyl-1,3-oxazolidin-2-ones as mGluR2 positive allosteric modulators: Hit-to lead and lead optimization. Bioorg Med Chem Lett19:2524-9 (2009) [PubMed] Article
Duplantier, AJ; Efremov, I; Candler, J; Doran, AC; Ganong, AH; Haas, JA; Hanks, AN; Kraus, KG; Lazzaro, JT; Lu, J; Maklad, N; McCarthy, SA; O'Sullivan, TJ; Rogers, BN; Siuciak, JA; Spracklin, DK; Zhang, L 3-Benzyl-1,3-oxazolidin-2-ones as mGluR2 positive allosteric modulators: Hit-to lead and lead optimization. Bioorg Med Chem Lett19:2524-9 (2009) [PubMed] Article