| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cathepsin D |
|---|
| Ligand | BDBM50305469 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_603779 (CHEMBL1042917) |
|---|
| IC50 | 17100±n/a nM |
|---|
| Citation |  Nowak, P; Cole, DC; Aulabaugh, A; Bard, J; Chopra, R; Cowling, R; Fan, KY; Hu, B; Jacobsen, S; Jani, M; Jin, G; Lo, MC; Malamas, MS; Manas, ES; Narasimhan, R; Reinhart, P; Robichaud, AJ; Stock, JR; Subrath, J; Svenson, K; Turner, J; Wagner, E; Zhou, P; Ellingboe, JW Discovery and initial optimization of 5,5'-disubstituted aminohydantoins as potent beta-secretase (BACE1) inhibitors. Bioorg Med Chem Lett20:632-5 (2010) [PubMed] Article Nowak, P; Cole, DC; Aulabaugh, A; Bard, J; Chopra, R; Cowling, R; Fan, KY; Hu, B; Jacobsen, S; Jani, M; Jin, G; Lo, MC; Malamas, MS; Manas, ES; Narasimhan, R; Reinhart, P; Robichaud, AJ; Stock, JR; Subrath, J; Svenson, K; Turner, J; Wagner, E; Zhou, P; Ellingboe, JW Discovery and initial optimization of 5,5'-disubstituted aminohydantoins as potent beta-secretase (BACE1) inhibitors. Bioorg Med Chem Lett20:632-5 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cathepsin D |
|---|
| Name: | Cathepsin D |
|---|
| Synonyms: | CATD_HUMAN | CPSD | CTSD | Cathepsin D [Precursor] | Cathepsin D heavy chain | Cathepsin D light chain | Cathepsin D precursor |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 44551.72 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Human proCathepsin D (SwissProt accession number P07339) was expressed in Sf9 cells, purified, and autoactivated. |
|---|
| Residue: | 412 |
|---|
| Sequence: | MQPSSLLPLALCLLAAPASALVRIPLHKFTSIRRTMSEVGGSVEDLIAKGPVSKYSQAVP
AVTEGPIPEVLKNYMDAQYYGEIGIGTPPQCFTVVFDTGSSNLWVPSIHCKLLDIACWIH
HKYNSDKSSTYVKNGTSFDIHYGSGSLSGYLSQDTVSVPCQSASSASALGGVKVERQVFG
EATKQPGITFIAAKFDGILGMAYPRISVNNVLPVFDNLMQQKLVDQNIFSFYLSRDPDAQ
PGGELMLGGTDSKYYKGSLSYLNVTRKAYWQVHLDQVEVASGLTLCKEGCEAIVDTGTSL
MVGPVDEVRELQKAIGAVPLIQGEYMIPCEKVSTLPAITLKLGGKGYKLSPEDYTLKVSQ
AGKTLCLSGFMGMDIPPPSGPLWILGDVFIGRYYTVFDRDNNRVGFAEAARL
|
|
|
|---|
| BDBM50305469 |
|---|
| n/a |
|---|
| Name | BDBM50305469 |
|---|
| Synonyms: | CHEMBL590830 | N-(3-(2-amino-4-cyclohexyl-1-methyl-5-oxo-4,5-dihydro-1H-imidazol-4-yl)phenyl)-3-methylfuran-2-carboxamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H26N4O3 |
|---|
| Mol. Mass. | 394.4668 |
|---|
| SMILES | CN1C(N)=NC(C2CCCCC2)(C1=O)c1cccc(NC(=O)c2occc2C)c1 |c:3| |
|---|
| Structure | 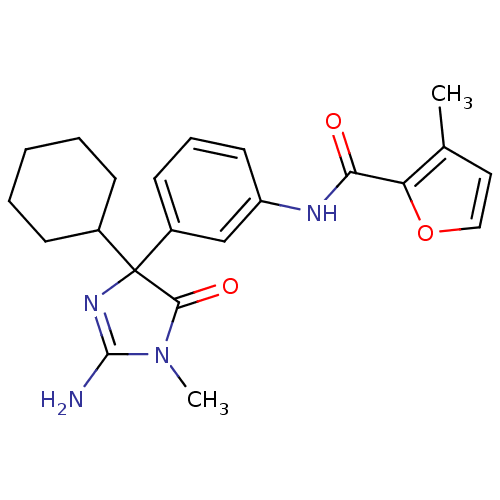
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Nowak, P; Cole, DC; Aulabaugh, A; Bard, J; Chopra, R; Cowling, R; Fan, KY; Hu, B; Jacobsen, S; Jani, M; Jin, G; Lo, MC; Malamas, MS; Manas, ES; Narasimhan, R; Reinhart, P; Robichaud, AJ; Stock, JR; Subrath, J; Svenson, K; Turner, J; Wagner, E; Zhou, P; Ellingboe, JW Discovery and initial optimization of 5,5'-disubstituted aminohydantoins as potent beta-secretase (BACE1) inhibitors. Bioorg Med Chem Lett20:632-5 (2010) [PubMed] Article
Nowak, P; Cole, DC; Aulabaugh, A; Bard, J; Chopra, R; Cowling, R; Fan, KY; Hu, B; Jacobsen, S; Jani, M; Jin, G; Lo, MC; Malamas, MS; Manas, ES; Narasimhan, R; Reinhart, P; Robichaud, AJ; Stock, JR; Subrath, J; Svenson, K; Turner, J; Wagner, E; Zhou, P; Ellingboe, JW Discovery and initial optimization of 5,5'-disubstituted aminohydantoins as potent beta-secretase (BACE1) inhibitors. Bioorg Med Chem Lett20:632-5 (2010) [PubMed] Article