

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Sodium-dependent dopamine transporter | ||
| Ligand | BDBM50334461 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_700838 (CHEMBL1646232) | ||
| Ki | 14±n/a nM | ||
| Citation |  Banister, SD; Moussa, IA; Beinat, C; Reynolds, AJ; Schiavini, P; Jorgensen, WT; Kassiou, M Trishomocubane as a scaffold for the development of selective dopamine transporter (DAT) ligands. Bioorg Med Chem Lett21:38-41 (2010) [PubMed] Article Banister, SD; Moussa, IA; Beinat, C; Reynolds, AJ; Schiavini, P; Jorgensen, WT; Kassiou, M Trishomocubane as a scaffold for the development of selective dopamine transporter (DAT) ligands. Bioorg Med Chem Lett21:38-41 (2010) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Sodium-dependent dopamine transporter | |||
| Name: | Sodium-dependent dopamine transporter | ||
| Synonyms: | DA transporter | DAT | DAT1 | Dopamine transporter (DAT) | Dopamine transporter protein (DAT) | Monoamine transporter | SC6A3_HUMAN | SLC6A3 | Sodium-dependent dopamine transporter | Sodium-dependent dopamine transporter (DAT) | Solute carrier family 6 member 3 | ||
| Type: | Multi-pass membrane protein | ||
| Mol. Mass.: | 68497.11 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q01959 | ||
| Residue: | 620 | ||
| Sequence: |
| ||
| BDBM50334461 | |||
| n/a | |||
| Name | BDBM50334461 | ||
| Synonyms: | 2-butyl-3-(4-chlorophenyl)-1-azabicyclo[2.2.2]oct-2-ene | CHEMBL1643898 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C17H22ClN | ||
| Mol. Mass. | 275.816 | ||
| SMILES | CCCCC1=C(C2CCN1CC2)c1ccc(Cl)cc1 |t:4,(.06,-16.55,;-1.41,-17.03,;-2.55,-16,;-4.01,-16.47,;-5.2,-15.49,;-5.2,-13.3,;-7.34,-12.86,;-6.01,-13.62,;-6.01,-15.16,;-7.35,-15.93,;-8.69,-15.16,;-8.66,-13.62,;-3.99,-12.34,;-2.55,-12.89,;-1.36,-11.93,;-1.59,-10.41,;-.39,-9.44,;-3.03,-9.85,;-4.23,-10.82,)| | ||
| Structure | 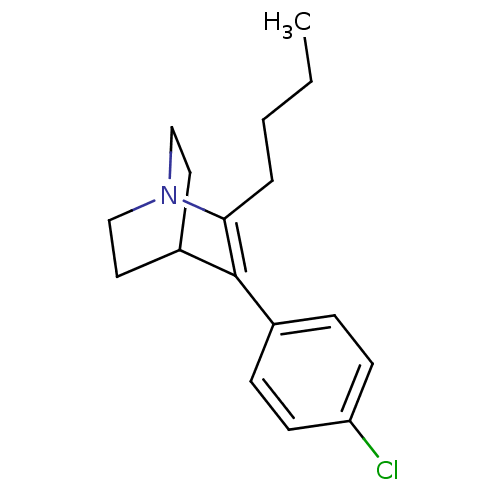
| ||