| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Induced myeloid leukemia cell differentiation protein Mcl-1 |
|---|
| Ligand | BDBM50339778 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_735636 (CHEMBL1693338) |
|---|
| IC50 | >1000±n/a nM |
|---|
| Citation |  Sleebs, BE; Czabotar, PE; Fairbrother, WJ; Fairlie, WD; Flygare, JA; Huang, DC; Kersten, WJ; Koehler, MF; Lessene, G; Lowes, K; Parisot, JP; Smith, BJ; Smith, ML; Souers, AJ; Street, IP; Yang, H; Baell, JB Quinazoline sulfonamides as dual binders of the proteins B-cell lymphoma 2 and B-cell lymphoma extra long with potent proapoptotic cell-based activity. J Med Chem54:1914-26 (2011) [PubMed] Article Sleebs, BE; Czabotar, PE; Fairbrother, WJ; Fairlie, WD; Flygare, JA; Huang, DC; Kersten, WJ; Koehler, MF; Lessene, G; Lowes, K; Parisot, JP; Smith, BJ; Smith, ML; Souers, AJ; Street, IP; Yang, H; Baell, JB Quinazoline sulfonamides as dual binders of the proteins B-cell lymphoma 2 and B-cell lymphoma extra long with potent proapoptotic cell-based activity. J Med Chem54:1914-26 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Induced myeloid leukemia cell differentiation protein Mcl-1 |
|---|
| Name: | Induced myeloid leukemia cell differentiation protein Mcl-1 |
|---|
| Synonyms: | BCL2L3 | Bcl-2-like protein 3 | Bcl-2-like protein 3 (Mcl-1) | Bcl-2-related protein EAT/mcl1 | Bcl2-L-3 | Induced myeloid leukemia cell differentiation protein (Mcl-1) | MCL1 | MCL1_HUMAN | Mcl-1 | Myeloid Cell factor-1 (Mcl-1) | Myeloid cell leukemia sequence 1 (BCL2-related) | mcl1/EAT |
|---|
| Type: | Membrane; Single-pass membrane protein |
|---|
| Mol. Mass.: | 37332.87 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q07820 |
|---|
| Residue: | 350 |
|---|
| Sequence: | MFGLKRNAVIGLNLYCGGAGLGAGSGGATRPGGRLLATEKEASARREIGGGEAGAVIGGS
AGASPPSTLTPDSRRVARPPPIGAEVPDVTATPARLLFFAPTRRAAPLEEMEAPAADAIM
SPEEELDGYEPEPLGKRPAVLPLLELVGESGNNTSTDGSLPSTPPPAEEEEDELYRQSLE
IISRYLREQATGAKDTKPMGRSGATSRKALETLRRVGDGVQRNHETAFQGMLRKLDIKNE
DDVKSLSRVMIHVFSDGVTNWGRIVTLISFGAFVAKHLKTINQESCIEPLAESITDVLVR
TKRDWLVKQRGWDGFVEFFHVEDLEGGIRNVLLAFAGVAGVGAGLAYLIR
|
|
|
|---|
| BDBM50339778 |
|---|
| n/a |
|---|
| Name | BDBM50339778 |
|---|
| Synonyms: | CHEMBL1689140 | N-{7-[4-(4'-Chloro-biphenyl-2-ylmethyl)piperazin-1-yl]quinazolin-4-yl}-4-((R)-3-morpholin-4-yl-1-phenylsulfanylmethyl-propylamino)-3-nitro-benzenesulfonamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C45H47ClN8O5S2 |
|---|
| Mol. Mass. | 879.488 |
|---|
| SMILES | [O-][N+](=O)c1cc(ccc1N[C@H](CCN1CCOCC1)CSc1ccccc1)S(=O)(=O)Nc1ncnc2cc(ccc12)N1CCN(Cc2ccccc2-c2ccc(Cl)cc2)CC1 |r| |
|---|
| Structure | 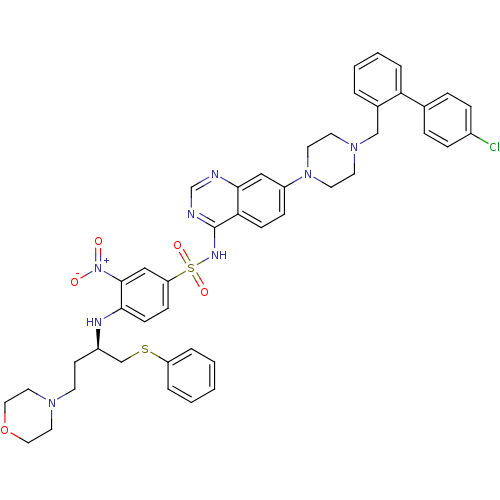
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Sleebs, BE; Czabotar, PE; Fairbrother, WJ; Fairlie, WD; Flygare, JA; Huang, DC; Kersten, WJ; Koehler, MF; Lessene, G; Lowes, K; Parisot, JP; Smith, BJ; Smith, ML; Souers, AJ; Street, IP; Yang, H; Baell, JB Quinazoline sulfonamides as dual binders of the proteins B-cell lymphoma 2 and B-cell lymphoma extra long with potent proapoptotic cell-based activity. J Med Chem54:1914-26 (2011) [PubMed] Article
Sleebs, BE; Czabotar, PE; Fairbrother, WJ; Fairlie, WD; Flygare, JA; Huang, DC; Kersten, WJ; Koehler, MF; Lessene, G; Lowes, K; Parisot, JP; Smith, BJ; Smith, ML; Souers, AJ; Street, IP; Yang, H; Baell, JB Quinazoline sulfonamides as dual binders of the proteins B-cell lymphoma 2 and B-cell lymphoma extra long with potent proapoptotic cell-based activity. J Med Chem54:1914-26 (2011) [PubMed] Article