| Reaction Details |
|---|
 | Report a problem with these data |
| Target | ATP-dependent translocase ABCB1 |
|---|
| Ligand | BDBM50369565 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_103350 (CHEMBL710068) |
|---|
| IC50 | 180±n/a nM |
|---|
| Citation |  Tiberghien, F; Kurome, T; Takesako, K; Didier, A; Wenandy, T; Loor, F Aureobasidins: structure-activity relationships for the inhibition of the human MDR1 P-glycoprotein ABC-transporter. J Med Chem43:2547-56 (2000) [PubMed] Tiberghien, F; Kurome, T; Takesako, K; Didier, A; Wenandy, T; Loor, F Aureobasidins: structure-activity relationships for the inhibition of the human MDR1 P-glycoprotein ABC-transporter. J Med Chem43:2547-56 (2000) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| ATP-dependent translocase ABCB1 |
|---|
| Name: | ATP-dependent translocase ABCB1 |
|---|
| Synonyms: | ABCB1 | MDR1 | MDR1_HUMAN | Multidrug Resistance Transporter MDR 1 | Multidrug resistance protein 1 | Multidrug resistance protein 1/Multidrug resistance associated protein 1 | P-glycoprotein (P-gp) | P-glycoprotein 1 | PGY1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 141503.50 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P08183 |
|---|
| Residue: | 1280 |
|---|
| Sequence: | MDLEGDRNGGAKKKNFFKLNNKSEKDKKEKKPTVSVFSMFRYSNWLDKLYMVVGTLAAII
HGAGLPLMMLVFGEMTDIFANAGNLEDLMSNITNRSDINDTGFFMNLEEDMTRYAYYYSG
IGAGVLVAAYIQVSFWCLAAGRQIHKIRKQFFHAIMRQEIGWFDVHDVGELNTRLTDDVS
KINEGIGDKIGMFFQSMATFFTGFIVGFTRGWKLTLVILAISPVLGLSAAVWAKILSSFT
DKELLAYAKAGAVAEEVLAAIRTVIAFGGQKKELERYNKNLEEAKRIGIKKAITANISIG
AAFLLIYASYALAFWYGTTLVLSGEYSIGQVLTVFFSVLIGAFSVGQASPSIEAFANARG
AAYEIFKIIDNKPSIDSYSKSGHKPDNIKGNLEFRNVHFSYPSRKEVKILKGLNLKVQSG
QTVALVGNSGCGKSTTVQLMQRLYDPTEGMVSVDGQDIRTINVRFLREIIGVVSQEPVLF
ATTIAENIRYGRENVTMDEIEKAVKEANAYDFIMKLPHKFDTLVGERGAQLSGGQKQRIA
IARALVRNPKILLLDEATSALDTESEAVVQVALDKARKGRTTIVIAHRLSTVRNADVIAG
FDDGVIVEKGNHDELMKEKGIYFKLVTMQTAGNEVELENAADESKSEIDALEMSSNDSRS
SLIRKRSTRRSVRGSQAQDRKLSTKEALDESIPPVSFWRIMKLNLTEWPYFVVGVFCAII
NGGLQPAFAIIFSKIIGVFTRIDDPETKRQNSNLFSLLFLALGIISFITFFLQGFTFGKA
GEILTKRLRYMVFRSMLRQDVSWFDDPKNTTGALTTRLANDAAQVKGAIGSRLAVITQNI
ANLGTGIIISFIYGWQLTLLLLAIVPIIAIAGVVEMKMLSGQALKDKKELEGSGKIATEA
IENFRTVVSLTQEQKFEHMYAQSLQVPYRNSLRKAHIFGITFSFTQAMMYFSYAGCFRFG
AYLVAHKLMSFEDVLLVFSAVVFGAMAVGQVSSFAPDYAKAKISAAHIIMIIEKTPLIDS
YSTEGLMPNTLEGNVTFGEVVFNYPTRPDIPVLQGLSLEVKKGQTLALVGSSGCGKSTVV
QLLERFYDPLAGKVLLDGKEIKRLNVQWLRAHLGIVSQEPILFDCSIAENIAYGDNSRVV
SQEEIVRAAKEANIHAFIESLPNKYSTKVGDKGTQLSGGQKQRIAIARALVRQPHILLLD
EATSALDTESEKVVQEALDKAREGRTCIVIAHRLSTIQNADLIVVFQNGRVKEHGTHQQL
LAQKGIYFSMVSVQAGTKRQ
|
|
|
|---|
| BDBM50369565 |
|---|
| n/a |
|---|
| Name | BDBM50369565 |
|---|
| Synonyms: | CHEMBL1790683 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C60H90N8O10 |
|---|
| Mol. Mass. | 1083.4042 |
|---|
| SMILES | [#6]-[#6]-[#6@H](-[#6])-[#6@@H]-1-[#7]-[#6](=O)-[#6@@H]-2-[#6]-[#6]-[#6]-[#7]-2-[#6](=O)-[#6@H](-[#6]-c2ccccc2)-[#7](-[#6])-[#6](=O)-[#6@H](-[#6]-c2ccccc2)-[#7]-[#6](=O)-[#6@H](-[#6](-[#6])-[#6])-[#7](-[#6])-[#6](=O)-[#6@H](-[#8]-[#6](=O)\[#6](-[#7](-[#6])-[#6](=O)-[#6@H](-[#6]-[#6](-[#6])-[#6])-[#7]-[#6](=O)-[#6@H](-[#6](-[#6])-[#6])-[#7](-[#6])-[#6]-1=O)=[#6](\[#6])-[#6])-[#6@@H](-[#6])-[#6]-[#6] |
|---|
| Structure | 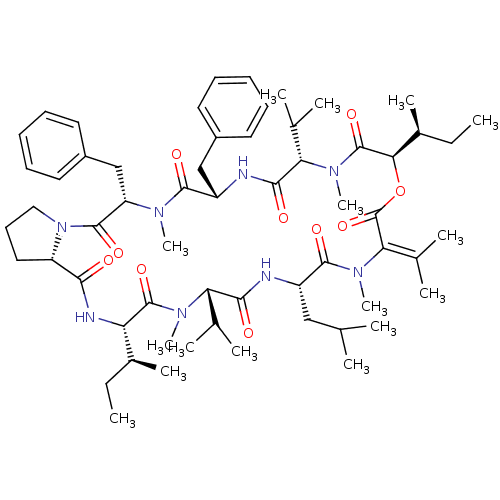
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Tiberghien, F; Kurome, T; Takesako, K; Didier, A; Wenandy, T; Loor, F Aureobasidins: structure-activity relationships for the inhibition of the human MDR1 P-glycoprotein ABC-transporter. J Med Chem43:2547-56 (2000) [PubMed]
Tiberghien, F; Kurome, T; Takesako, K; Didier, A; Wenandy, T; Loor, F Aureobasidins: structure-activity relationships for the inhibition of the human MDR1 P-glycoprotein ABC-transporter. J Med Chem43:2547-56 (2000) [PubMed]