| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Induced myeloid leukemia cell differentiation protein Mcl-1 |
|---|
| Ligand | BDBM50393839 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_854897 (CHEMBL2162634) |
|---|
| IC50 | 37±n/a nM |
|---|
| Citation |  Zhou, H; Aguilar, A; Chen, J; Bai, L; Liu, L; Meagher, JL; Yang, CY; McEachern, D; Cong, X; Stuckey, JA; Wang, S Structure-based design of potent Bcl-2/Bcl-xL inhibitors with strong in vivo antitumor activity. J Med Chem55:6149-61 (2012) [PubMed] Article Zhou, H; Aguilar, A; Chen, J; Bai, L; Liu, L; Meagher, JL; Yang, CY; McEachern, D; Cong, X; Stuckey, JA; Wang, S Structure-based design of potent Bcl-2/Bcl-xL inhibitors with strong in vivo antitumor activity. J Med Chem55:6149-61 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Induced myeloid leukemia cell differentiation protein Mcl-1 |
|---|
| Name: | Induced myeloid leukemia cell differentiation protein Mcl-1 |
|---|
| Synonyms: | BCL2L3 | Bcl-2-like protein 3 | Bcl-2-like protein 3 (Mcl-1) | Bcl-2-related protein EAT/mcl1 | Bcl2-L-3 | Induced myeloid leukemia cell differentiation protein (Mcl-1) | MCL1 | MCL1_HUMAN | Mcl-1 | Myeloid Cell factor-1 (Mcl-1) | Myeloid cell leukemia sequence 1 (BCL2-related) | mcl1/EAT |
|---|
| Type: | Membrane; Single-pass membrane protein |
|---|
| Mol. Mass.: | 37332.87 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q07820 |
|---|
| Residue: | 350 |
|---|
| Sequence: | MFGLKRNAVIGLNLYCGGAGLGAGSGGATRPGGRLLATEKEASARREIGGGEAGAVIGGS
AGASPPSTLTPDSRRVARPPPIGAEVPDVTATPARLLFFAPTRRAAPLEEMEAPAADAIM
SPEEELDGYEPEPLGKRPAVLPLLELVGESGNNTSTDGSLPSTPPPAEEEEDELYRQSLE
IISRYLREQATGAKDTKPMGRSGATSRKALETLRRVGDGVQRNHETAFQGMLRKLDIKNE
DDVKSLSRVMIHVFSDGVTNWGRIVTLISFGAFVAKHLKTINQESCIEPLAESITDVLVR
TKRDWLVKQRGWDGFVEFFHVEDLEGGIRNVLLAFAGVAGVGAGLAYLIR
|
|
|
|---|
| BDBM50393839 |
|---|
| n/a |
|---|
| Name | BDBM50393839 |
|---|
| Synonyms: | CHEMBL2159744 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C116H190N36O35S |
|---|
| Mol. Mass. | 2681.035 |
|---|
| SMILES | [#6]-[#6](-[#6])-[#6]-[#6@H](-[#7]-[#6](=O)-[#6@@H](-[#7])-[#6]-[#6]-[#6](-[#8])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6](-[#8])=O)-[#6](=O)-[#7]-[#6@@H](-[#6](-[#6])-[#6])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6](-[#8])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#16])-[#6](=O)-[#7]-[#6@@H](-[#6])-[#6](=O)-[#7]-[#6@@H](-[#6@@H](-[#6])-[#8])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6](-[#7])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6](-[#6])-[#6])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#6](=O)-[#7]-[#6@@H](-[#6]-c1ccccc1)-[#6](=O)-[#7]-[#6]-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6](-[#8])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]-[#6]-[#7])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6](-[#6])-[#6])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6](-[#7])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-c1ccccc1)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6](-[#7])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]-[#6]-[#7])-[#6](-[#8])=O |r| |
|---|
| Structure | 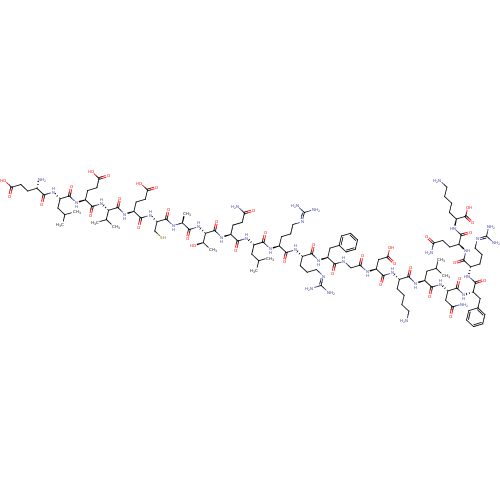
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Zhou, H; Aguilar, A; Chen, J; Bai, L; Liu, L; Meagher, JL; Yang, CY; McEachern, D; Cong, X; Stuckey, JA; Wang, S Structure-based design of potent Bcl-2/Bcl-xL inhibitors with strong in vivo antitumor activity. J Med Chem55:6149-61 (2012) [PubMed] Article
Zhou, H; Aguilar, A; Chen, J; Bai, L; Liu, L; Meagher, JL; Yang, CY; McEachern, D; Cong, X; Stuckey, JA; Wang, S Structure-based design of potent Bcl-2/Bcl-xL inhibitors with strong in vivo antitumor activity. J Med Chem55:6149-61 (2012) [PubMed] Article