| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Macrophage-stimulating protein receptor |
|---|
| Ligand | BDBM50427140 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_940647 (CHEMBL2327589) |
|---|
| IC50 | 912±n/a nM |
|---|
| Citation |  Northrup, AB; Katcher, MH; Altman, MD; Chenard, M; Daniels, MH; Deshmukh, SV; Falcone, D; Guerin, DJ; Hatch, H; Li, C; Lu, W; Lutterbach, B; Allison, TJ; Patel, SB; Reilly, JF; Reutershan, M; Rickert, KW; Rosenstein, C; Soisson, SM; Szewczak, AA; Walker, D; Wilson, K; Young, JR; Pan, BS; Dinsmore, CJ Discovery of 1-[3-(1-methyl-1H-pyrazol-4-yl)-5-oxo-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl]-N-(pyridin-2-ylmethyl)methanesulfonamide (MK-8033): A Specific c-Met/Ron dual kinase inhibitor with preferential affinity for the activated state of c-Met. J Med Chem56:2294-310 (2013) [PubMed] Article Northrup, AB; Katcher, MH; Altman, MD; Chenard, M; Daniels, MH; Deshmukh, SV; Falcone, D; Guerin, DJ; Hatch, H; Li, C; Lu, W; Lutterbach, B; Allison, TJ; Patel, SB; Reilly, JF; Reutershan, M; Rickert, KW; Rosenstein, C; Soisson, SM; Szewczak, AA; Walker, D; Wilson, K; Young, JR; Pan, BS; Dinsmore, CJ Discovery of 1-[3-(1-methyl-1H-pyrazol-4-yl)-5-oxo-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl]-N-(pyridin-2-ylmethyl)methanesulfonamide (MK-8033): A Specific c-Met/Ron dual kinase inhibitor with preferential affinity for the activated state of c-Met. J Med Chem56:2294-310 (2013) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Macrophage-stimulating protein receptor |
|---|
| Name: | Macrophage-stimulating protein receptor |
|---|
| Synonyms: | 2.7.10.1 | CD_antigen=CD136 | CDw136 | MSP receptor | MST1R | Macrophage-stimulating protein receptor (MST1R) | Macrophage-stimulating protein receptor alpha chain | Macrophage-stimulating protein receptor beta chain | PTK8 | Protein-tyrosine kinase 8 | RON | RON_HUMAN | Tyrosine kinase receptor ron | p185-Ron |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 152270.76 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q04912 |
|---|
| Residue: | 1400 |
|---|
| Sequence: | MELLPPLPQSFLLLLLLPAKPAAGEDWQCPRTPYAASRDFDVKYVVPSFSAGGLVQAMVT
YEGDRNESAVFVAIRNRLHVLGPDLKSVQSLATGPAGDPGCQTCAACGPGPHGPPGDTDT
KVLVLDPALPALVSCGSSLQGRCFLHDLEPQGTAVHLAAPACLFSAHHNRPDDCPDCVAS
PLGTRVTVVEQGQASYFYVASSLDAAVAASFSPRSVSIRRLKADASGFAPGFVALSVLPK
HLVSYSIEYVHSFHTGAFVYFLTVQPASVTDDPSALHTRLARLSATEPELGDYRELVLDC
RFAPKRRRRGAPEGGQPYPVLRVAHSAPVGAQLATELSIAEGQEVLFGVFVTGKDGGPGV
GPNSVVCAFPIDLLDTLIDEGVERCCESPVHPGLRRGLDFFQSPSFCPNPPGLEALSPNT
SCRHFPLLVSSSFSRVDLFNGLLGPVQVTALYVTRLDNVTVAHMGTMDGRILQVELVRSL
NYLLYVSNFSLGDSGQPVQRDVSRLGDHLLFASGDQVFQVPIQGPGCRHFLTCGRCLRAW
HFMGCGWCGNMCGQQKECPGSWQQDHCPPKLTEFHPHSGPLRGSTRLTLCGSNFYLHPSG
LVPEGTHQVTVGQSPCRPLPKDSSKLRPVPRKDFVEEFECELEPLGTQAVGPTNVSLTVT
NMPPGKHFRVDGTSVLRGFSFMEPVLIAVQPLFGPRAGGTCLTLEGQSLSVGTSRAVLVN
GTECLLARVSEGQLLCATPPGATVASVPLSLQVGGAQVPGSWTFQYREDPVVLSISPNCG
YINSHITICGQHLTSAWHLVLSFHDGLRAVESRCERQLPEQQLCRLPEYVVRDPQGWVAG
NLSARGDGAAGFTLPGFRFLPPPHPPSANLVPLKPEEHAIKFEYIGLGAVADCVGINVTV
GGESCQHEFRGDMVVCPLPPSLQLGQDGAPLQVCVDGECHILGRVVRPGPDGVPQSTLLG
ILLPLLLLVAALATALVFSYWWRRKQLVLPPNLNDLASLDQTAGATPLPILYSGSDYRSG
LALPAIDGLDSTTCVHGASFSDSEDESCVPLLRKESIQLRDLDSALLAEVKDVLIPHERV
VTHSDRVIGKGHFGVVYHGEYIDQAQNRIQCAIKSLSRITEMQQVEAFLREGLLMRGLNH
PNVLALIGIMLPPEGLPHVLLPYMCHGDLLQFIRSPQRNPTVKDLISFGLQVARGMEYLA
EQKFVHRDLAARNCMLDESFTVKVADFGLARDILDREYYSVQQHRHARLPVKWMALESLQ
TYRFTTKSDVWSFGVLLWELLTRGAPPYRHIDPFDLTHFLAQGRRLPQPEYCPDSLYQVM
QQCWEADPAVRPTFRVLVGEVEQIVSALLGDHYVQLPATYMNLGPSTSHEMNVRPEQPQF
SPMPGNVRRPRPLSEPPRPT
|
|
|
|---|
| BDBM50427140 |
|---|
| n/a |
|---|
| Name | BDBM50427140 |
|---|
| Synonyms: | CHEMBL2324116 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H18N4O2 |
|---|
| Mol. Mass. | 358.3932 |
|---|
| SMILES | CC(C(N)=O)c1ccc2ccc3ncc(cc3c(=O)c2c1)-c1cnn(C)c1 |
|---|
| Structure | 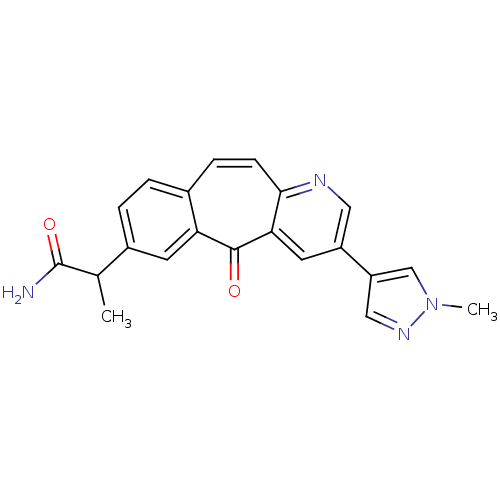
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Northrup, AB; Katcher, MH; Altman, MD; Chenard, M; Daniels, MH; Deshmukh, SV; Falcone, D; Guerin, DJ; Hatch, H; Li, C; Lu, W; Lutterbach, B; Allison, TJ; Patel, SB; Reilly, JF; Reutershan, M; Rickert, KW; Rosenstein, C; Soisson, SM; Szewczak, AA; Walker, D; Wilson, K; Young, JR; Pan, BS; Dinsmore, CJ Discovery of 1-[3-(1-methyl-1H-pyrazol-4-yl)-5-oxo-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl]-N-(pyridin-2-ylmethyl)methanesulfonamide (MK-8033): A Specific c-Met/Ron dual kinase inhibitor with preferential affinity for the activated state of c-Met. J Med Chem56:2294-310 (2013) [PubMed] Article
Northrup, AB; Katcher, MH; Altman, MD; Chenard, M; Daniels, MH; Deshmukh, SV; Falcone, D; Guerin, DJ; Hatch, H; Li, C; Lu, W; Lutterbach, B; Allison, TJ; Patel, SB; Reilly, JF; Reutershan, M; Rickert, KW; Rosenstein, C; Soisson, SM; Szewczak, AA; Walker, D; Wilson, K; Young, JR; Pan, BS; Dinsmore, CJ Discovery of 1-[3-(1-methyl-1H-pyrazol-4-yl)-5-oxo-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl]-N-(pyridin-2-ylmethyl)methanesulfonamide (MK-8033): A Specific c-Met/Ron dual kinase inhibitor with preferential affinity for the activated state of c-Met. J Med Chem56:2294-310 (2013) [PubMed] Article