

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Bifunctional epoxide hydrolase 2 | ||
| Ligand | BDBM50435134 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_963496 (CHEMBL2394377) | ||
| IC50 | 936±n/a nM | ||
| Citation |  North, EJ; Scherman, MS; Bruhn, DF; Scarborough, JS; Maddox, MM; Jones, V; Grzegorzewicz, A; Yang, L; Hess, T; Morisseau, C; Jackson, M; McNeil, MR; Lee, RE Design, synthesis and anti-tuberculosis activity of 1-adamantyl-3-heteroaryl ureas with improved in vitro pharmacokinetic properties. Bioorg Med Chem21:2587-99 (2013) [PubMed] Article North, EJ; Scherman, MS; Bruhn, DF; Scarborough, JS; Maddox, MM; Jones, V; Grzegorzewicz, A; Yang, L; Hess, T; Morisseau, C; Jackson, M; McNeil, MR; Lee, RE Design, synthesis and anti-tuberculosis activity of 1-adamantyl-3-heteroaryl ureas with improved in vitro pharmacokinetic properties. Bioorg Med Chem21:2587-99 (2013) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Bifunctional epoxide hydrolase 2 | |||
| Name: | Bifunctional epoxide hydrolase 2 | ||
| Synonyms: | Cytosolic epoxide hydrolase 2 | EBifunctional epoxide hydrolase 2 | EPHX2 | Epoxide hydratase | HYES_HUMAN | Lipid-phosphate phosphatase | Soluble epoxide hydrolase (sEH) | epoxide hydrolase 2, cytoplasmic | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 62613.07 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P34913 | ||
| Residue: | 555 | ||
| Sequence: |
| ||
| BDBM50435134 | |||
| n/a | |||
| Name | BDBM50435134 | ||
| Synonyms: | CHEMBL2392718 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C18H28N4O | ||
| Mol. Mass. | 316.4411 | ||
| SMILES | CCCn1nc(NC(=O)NC2C3CC4CC(C3)CC2C4)cc1C |TLB:9:10:12:16.15.14,THB:10:11:14:19.18.17,10:18:12.11.16:14,17:18:12:16.15.14,17:15:12:19.10.18,(29.77,-16.54,;29.31,-18.01,;27.8,-18.34,;27.34,-19.81,;25.88,-20.3,;25.89,-21.85,;24.56,-22.63,;23.23,-21.86,;23.23,-20.32,;21.9,-22.63,;20.57,-21.85,;20.07,-23.28,;20.56,-24.76,;19.23,-23.62,;17.75,-23.71,;17.2,-22.38,;18.49,-23.36,;17.71,-20.96,;19.24,-20.9,;19.66,-22.27,;27.36,-22.3,;28.25,-21.05,;29.79,-21.04,)| | ||
| Structure | 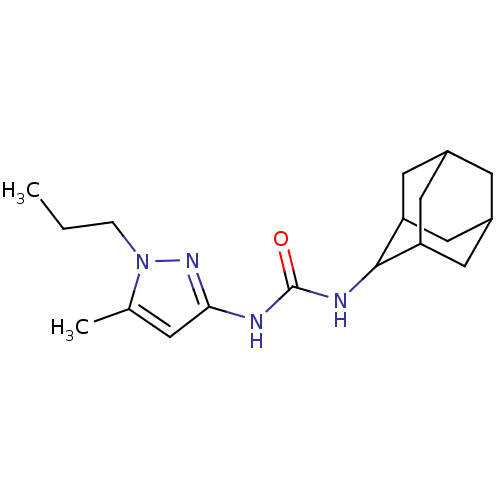
| ||