

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Poly [ADP-ribose] polymerase 2 | ||
| Ligand | BDBM50444584 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1282121 (CHEMBL3102005) | ||
| IC50 | >170000±n/a nM | ||
| Citation |  Huang, H; Guzman-Perez, A; Acquaviva, L; Berry, V; Bregman, H; Dovey, J; Gunaydin, H; Huang, X; Huang, L; Saffran, D; Serafino, R; Schneider, S; Wilson, C; DiMauro, EF Structure-based design of 2-aminopyridine oxazolidinones as potent and selective tankyrase inhibitors. ACS Med Chem Lett4:1218-23 (2013) [PubMed] Article Huang, H; Guzman-Perez, A; Acquaviva, L; Berry, V; Bregman, H; Dovey, J; Gunaydin, H; Huang, X; Huang, L; Saffran, D; Serafino, R; Schneider, S; Wilson, C; DiMauro, EF Structure-based design of 2-aminopyridine oxazolidinones as potent and selective tankyrase inhibitors. ACS Med Chem Lett4:1218-23 (2013) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Poly [ADP-ribose] polymerase 2 | |||
| Name: | Poly [ADP-ribose] polymerase 2 | ||
| Synonyms: | (ARTD2 or PARP2) | ADPRT2 | ADPRTL2 | PARP2 | PARP2_HUMAN | Poly [ADP-ribose] polymerase 2 (PARP-2) | Poly [ADP-ribose] polymerase 2 (PARP2) | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 66225.70 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q9UGN5 | ||
| Residue: | 583 | ||
| Sequence: |
| ||
| BDBM50444584 | |||
| n/a | |||
| Name | BDBM50444584 | ||
| Synonyms: | CHEMBL3099718 | US9340549, 95 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H26N4O2 | ||
| Mol. Mass. | 390.4781 | ||
| SMILES | CC1(C)OC(=O)N([C@H]1c1ccccc1)[C@H]1CC[C@@H](CC1)c1cnc(N)c(c1)C#N |r,wU:17.22,7.8,wD:14.15,(26.78,-18.45,;28.12,-19.21,;26.8,-19.99,;29.01,-17.95,;30.48,-18.41,;31.72,-17.5,;30.5,-19.95,;29.04,-20.44,;28.31,-21.8,;26.77,-21.84,;26.04,-23.2,;26.85,-24.51,;28.4,-24.45,;29.12,-23.1,;31.75,-20.85,;31.74,-22.4,;33.08,-23.17,;34.42,-22.4,;34.41,-20.85,;33.08,-20.08,;35.74,-23.16,;37.07,-22.39,;38.4,-23.15,;38.41,-24.7,;39.75,-25.47,;37.08,-25.47,;35.74,-24.7,;37.09,-27.01,;37.09,-28.55,)| | ||
| Structure | 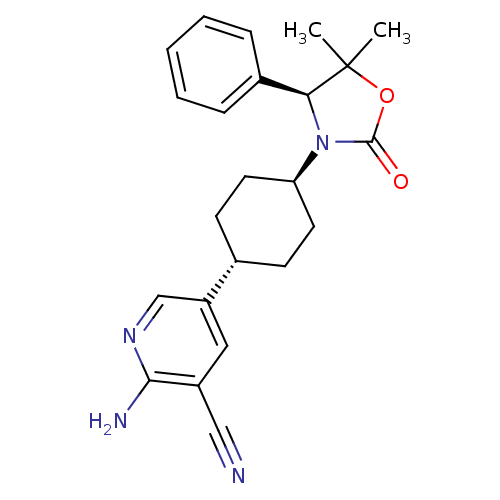
| ||