

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Poly [ADP-ribose] polymerase 1 | ||
| Ligand | BDBM50444583 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1282122 (CHEMBL3102006) | ||
| IC50 | >85000±n/a nM | ||
| Citation |  Huang, H; Guzman-Perez, A; Acquaviva, L; Berry, V; Bregman, H; Dovey, J; Gunaydin, H; Huang, X; Huang, L; Saffran, D; Serafino, R; Schneider, S; Wilson, C; DiMauro, EF Structure-based design of 2-aminopyridine oxazolidinones as potent and selective tankyrase inhibitors. ACS Med Chem Lett4:1218-23 (2013) [PubMed] Article Huang, H; Guzman-Perez, A; Acquaviva, L; Berry, V; Bregman, H; Dovey, J; Gunaydin, H; Huang, X; Huang, L; Saffran, D; Serafino, R; Schneider, S; Wilson, C; DiMauro, EF Structure-based design of 2-aminopyridine oxazolidinones as potent and selective tankyrase inhibitors. ACS Med Chem Lett4:1218-23 (2013) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Poly [ADP-ribose] polymerase 1 | |||
| Name: | Poly [ADP-ribose] polymerase 1 | ||
| Synonyms: | (ARTD1 or PARP1) | 2.4.2.- | 2.4.2.30 | ADP-ribosyltransferase diphtheria toxin-like 1 | ADPRT | ADPRT 1 | ARTD1 | DNA ADP-ribosyltransferase PARP1 | Human diphtheria toxin-like ADP-ribosyltransferase (ARTD1 or PARP1) | NAD(+) ADP-ribosyltransferase 1 | NT-PARP-1 | PARP-1 | PARP1 | PARP1_HUMAN | PPOL | Poly [ADP-ribose] polymerase (PARP) | Poly [ADP-ribose] polymerase 1 (PARP) | Poly [ADP-ribose] polymerase 1 (PARP-1) | Poly [ADP-ribose] polymerase 1 (PARP1) | Poly [ADP-ribose] polymerase 1, 24-kDa form | Poly [ADP-ribose] polymerase 1, 28-kDa form | Poly [ADP-ribose] polymerase 1, 89-kDa form | Poly [ADP-ribose] polymerase 1, processed C-terminus | Poly [ADP-ribose] polymerase 1, processed N-terminus | Poly [ADP-ribose] polymerase-1 | Poly(ADP-ribose) polymerase 1 (PARP1) | Poly(ADP-ribose) polymerase-1 (ARTD1/PARP1) | Poly[ADP-ribose] synthase 1 | Protein poly-ADP-ribosyltransferase PARP1 | Synonyms=ADPRT | ||
| Type: | n/a | ||
| Mol. Mass.: | 113114.22 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P09874 | ||
| Residue: | 1014 | ||
| Sequence: |
| ||
| BDBM50444583 | |||
| n/a | |||
| Name | BDBM50444583 | ||
| Synonyms: | CHEMBL3099716 | US9340549, 74 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C26H29N5O2 | ||
| Mol. Mass. | 443.5408 | ||
| SMILES | CC1(C)OC(=O)N([C@H]1c1ccccc1)[C@H]1CC[C@@H](CC1)c1cnc(N)c(c1)-c1ncccn1 |r,wU:17.22,7.8,wD:14.15,(49.17,-3.5,;50.51,-4.26,;49.19,-5.04,;51.4,-3,;52.87,-3.46,;54.11,-2.55,;52.89,-5,;51.43,-5.5,;50.7,-6.85,;49.16,-6.89,;48.43,-8.25,;49.24,-9.56,;50.79,-9.51,;51.51,-8.15,;54.14,-5.9,;54.14,-7.45,;55.47,-8.22,;56.81,-7.45,;56.8,-5.9,;55.47,-5.13,;58.14,-8.21,;59.46,-7.44,;60.8,-8.2,;60.8,-9.75,;62.14,-10.52,;59.47,-10.52,;58.14,-9.76,;59.47,-12.05,;58.14,-12.83,;58.14,-14.36,;59.47,-15.13,;60.81,-14.35,;60.8,-12.82,)| | ||
| Structure | 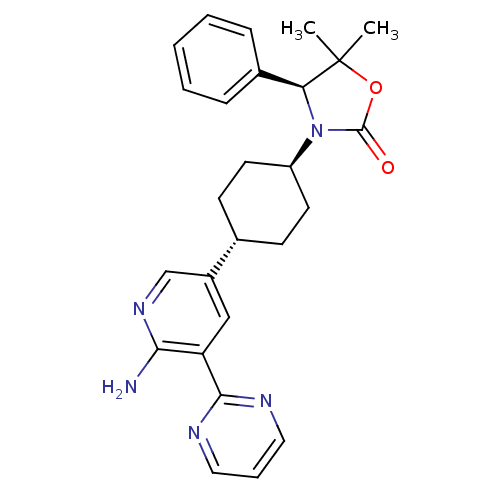
| ||