| Citation |  Zhang, N; Zhang, X; Zhu, J; Turpoff, A; Chen, G; Morrill, C; Huang, S; Lennox, W; Kakarla, R; Liu, R; Li, C; Ren, H; Almstead, N; Venkatraman, S; Njoroge, FG; Gu, Z; Clausen, V; Graci, J; Jung, SP; Zheng, Y; Colacino, JM; Lahser, F; Sheedy, J; Mollin, A; Weetall, M; Nomeir, A; Karp, GM Structure-activity relationship (SAR) optimization of 6-(indol-2-yl)pyridine-3-sulfonamides: identification of potent, selective, and orally bioavailable small molecules targeting hepatitis C (HCV) NS4B. J Med Chem57:2121-35 (2014) [PubMed] Article Zhang, N; Zhang, X; Zhu, J; Turpoff, A; Chen, G; Morrill, C; Huang, S; Lennox, W; Kakarla, R; Liu, R; Li, C; Ren, H; Almstead, N; Venkatraman, S; Njoroge, FG; Gu, Z; Clausen, V; Graci, J; Jung, SP; Zheng, Y; Colacino, JM; Lahser, F; Sheedy, J; Mollin, A; Weetall, M; Nomeir, A; Karp, GM Structure-activity relationship (SAR) optimization of 6-(indol-2-yl)pyridine-3-sulfonamides: identification of potent, selective, and orally bioavailable small molecules targeting hepatitis C (HCV) NS4B. J Med Chem57:2121-35 (2014) [PubMed] Article |
|---|
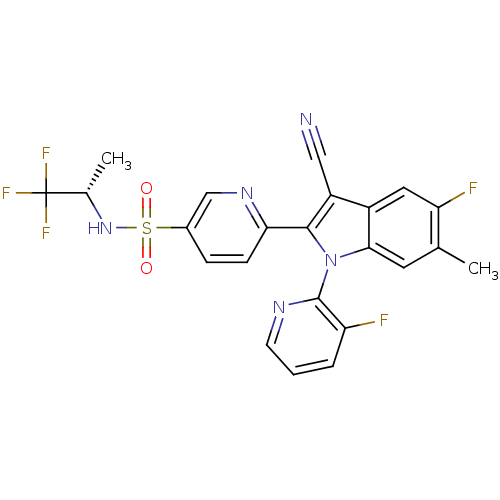
| SMILES | C[C@H](NS(=O)(=O)c1ccc(nc1)-c1c(C#N)c2cc(F)c(C)cc2n1-c1ncccc1F)C(F)(F)F |r,wD:1.0,(62.21,-19.89,;61.43,-18.56,;59.89,-18.57,;59.13,-19.9,;59.14,-21.44,;60.47,-20.67,;57.6,-19.91,;56.81,-18.58,;55.27,-18.59,;54.52,-19.93,;55.29,-21.26,;56.83,-21.25,;52.98,-19.94,;52.07,-18.7,;52.53,-17.23,;53,-15.76,;50.6,-19.18,;49.27,-18.42,;47.94,-19.19,;46.6,-18.42,;47.94,-20.73,;46.6,-21.5,;49.27,-21.5,;50.61,-20.72,;52.08,-21.19,;52.85,-22.52,;54.38,-22.51,;55.15,-23.83,;54.38,-25.17,;52.85,-25.17,;52.08,-23.84,;50.54,-23.84,;62.2,-17.22,;63.74,-17.21,;61.42,-15.89,;62.95,-15.87,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Zhang, N; Zhang, X; Zhu, J; Turpoff, A; Chen, G; Morrill, C; Huang, S; Lennox, W; Kakarla, R; Liu, R; Li, C; Ren, H; Almstead, N; Venkatraman, S; Njoroge, FG; Gu, Z; Clausen, V; Graci, J; Jung, SP; Zheng, Y; Colacino, JM; Lahser, F; Sheedy, J; Mollin, A; Weetall, M; Nomeir, A; Karp, GM Structure-activity relationship (SAR) optimization of 6-(indol-2-yl)pyridine-3-sulfonamides: identification of potent, selective, and orally bioavailable small molecules targeting hepatitis C (HCV) NS4B. J Med Chem57:2121-35 (2014) [PubMed] Article
Zhang, N; Zhang, X; Zhu, J; Turpoff, A; Chen, G; Morrill, C; Huang, S; Lennox, W; Kakarla, R; Liu, R; Li, C; Ren, H; Almstead, N; Venkatraman, S; Njoroge, FG; Gu, Z; Clausen, V; Graci, J; Jung, SP; Zheng, Y; Colacino, JM; Lahser, F; Sheedy, J; Mollin, A; Weetall, M; Nomeir, A; Karp, GM Structure-activity relationship (SAR) optimization of 6-(indol-2-yl)pyridine-3-sulfonamides: identification of potent, selective, and orally bioavailable small molecules targeting hepatitis C (HCV) NS4B. J Med Chem57:2121-35 (2014) [PubMed] Article