

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Potassium voltage-gated channel subfamily A member 5 | ||
| Ligand | BDBM50022113 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1367553 | ||
| IC50 | 88±n/a nM | ||
| Citation |  Johnson, JA; Xu, N; Jeon, Y; Finlay, HJ; Kover, A; Conder, ML; Sun, H; Li, D; Levesque, P; Hsueh, MM; Harper, TW; Wexler, RR; Lloyd, J Design, synthesis and evaluation of phenethylaminoheterocycles as K(v)1.5 inhibitors. Bioorg Med Chem Lett24:3018-22 (2014) [PubMed] Article Johnson, JA; Xu, N; Jeon, Y; Finlay, HJ; Kover, A; Conder, ML; Sun, H; Li, D; Levesque, P; Hsueh, MM; Harper, TW; Wexler, RR; Lloyd, J Design, synthesis and evaluation of phenethylaminoheterocycles as K(v)1.5 inhibitors. Bioorg Med Chem Lett24:3018-22 (2014) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Potassium voltage-gated channel subfamily A member 5 | |||
| Name: | Potassium voltage-gated channel subfamily A member 5 | ||
| Synonyms: | HK2 | HPCN1 | KCNA5 | KCNA5_HUMAN | Potassium voltage-gated channel subfamily A member 5 | Voltage-gated potassium channel | Voltage-gated potassium channel subunit Kv1.5 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 67222.47 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P22460 | ||
| Residue: | 613 | ||
| Sequence: |
| ||
| BDBM50022113 | |||
| n/a | |||
| Name | BDBM50022113 | ||
| Synonyms: | CHEMBL3298231 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C25H32N4O3 | ||
| Mol. Mass. | 436.5466 | ||
| SMILES | CCNC(=O)O[C@H]1CC[C@@](CNc2nn(C)c3cccc(OC)c23)(CC1)c1ccccc1 |r,wU:9.9,6.5,(16.91,-27.75,;15.56,-28.49,;14.24,-27.7,;12.89,-28.44,;12.86,-29.98,;11.58,-27.64,;11.61,-26.1,;10.29,-25.3,;10.33,-23.76,;11.68,-23.02,;13.02,-22.27,;14.34,-23.07,;15.69,-22.32,;15.68,-20.79,;17.13,-20.3,;17.6,-18.82,;18.04,-21.53,;19.58,-21.69,;20.22,-23.1,;19.31,-24.35,;17.78,-24.19,;16.88,-25.44,;17.51,-26.85,;17.15,-22.78,;13,-23.81,;12.96,-25.35,;10.36,-22.23,;10.4,-20.69,;9.08,-19.9,;7.73,-20.64,;7.71,-22.19,;9.02,-22.98,)| | ||
| Structure | 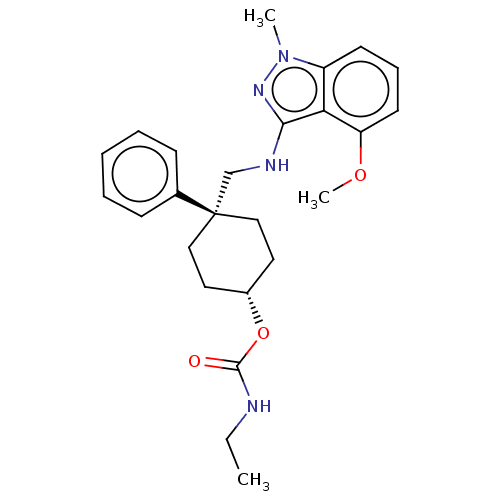
| ||