

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Poly [ADP-ribose] polymerase tankyrase-1 | ||
| Ligand | BDBM50082111 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1475151 (CHEMBL3424869) | ||
| IC50 | 18000±n/a nM | ||
| Citation |  Nencini, A; Pratelli, C; Quinn, JM; Salerno, M; Tunici, P; De Robertis, A; Valensin, S; Mennillo, F; Rossi, M; Bakker, A; Benicchi, T; Cappelli, F; Turlizzi, E; Nibbio, M; Caradonna, NP; Zanelli, U; Andreini, M; Magnani, M; Varrone, M Structure-activity relationship and properties optimization of a series of quinazoline-2,4-diones as inhibitors of the canonical Wnt pathway. Eur J Med Chem95:526-45 (2015) [PubMed] Article Nencini, A; Pratelli, C; Quinn, JM; Salerno, M; Tunici, P; De Robertis, A; Valensin, S; Mennillo, F; Rossi, M; Bakker, A; Benicchi, T; Cappelli, F; Turlizzi, E; Nibbio, M; Caradonna, NP; Zanelli, U; Andreini, M; Magnani, M; Varrone, M Structure-activity relationship and properties optimization of a series of quinazoline-2,4-diones as inhibitors of the canonical Wnt pathway. Eur J Med Chem95:526-45 (2015) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Poly [ADP-ribose] polymerase tankyrase-1 | |||
| Name: | Poly [ADP-ribose] polymerase tankyrase-1 | ||
| Synonyms: | (ARTD5 or PARP5a) | PARP5A | PARPL | Poly [ADP-ribose] polymerase 5 (PARP5) | Poly [ADP-ribose] polymerase tankyrase-1 | TIN1 | TINF1 | TNKS | TNKS1 | TNKS1_HUMAN | Tankyrase 1 | Tankyrase 1/2 | Tankyrase-1 | Tankyrase-1 (TNKS-1) | Tankyrase-1 (TNKS1) | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 142058.03 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | O95271 | ||
| Residue: | 1327 | ||
| Sequence: |
| ||
| BDBM50082111 | |||
| n/a | |||
| Name | BDBM50082111 | ||
| Synonyms: | CHEMBL3422630 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C25H34N4O6 | ||
| Mol. Mass. | 486.5607 | ||
| SMILES | COCC(=O)N1CCN(CC1)C(=O)[C@H]1CC[C@H](Cn2c(=O)n(C)c3ccc(OC)cc3c2=O)CC1 |r,wU:16.17,wD:13.13,(-16.01,5.07,;-16.01,3.84,;-14.68,3.07,;-14.68,1.53,;-15.75,.91,;-13.35,.75,;-13.35,-.79,;-12.02,-1.56,;-10.68,-.79,;-10.68,.75,;-12.02,1.52,;-9.35,-1.56,;-9.34,-2.79,;-8.01,-.78,;-6.68,-1.55,;-5.34,-.78,;-5.35,.76,;-4.02,1.54,;-2.68,.77,;-2.68,-.77,;-3.75,-1.39,;-1.33,-1.54,;-1.33,-2.77,;,-.77,;1.33,-1.54,;2.66,-.77,;2.66,.77,;4,1.54,;5.06,.92,;1.33,1.54,;,.77,;-1.33,1.54,;-1.33,2.77,;-6.69,1.53,;-8.02,.76,)| | ||
| Structure | 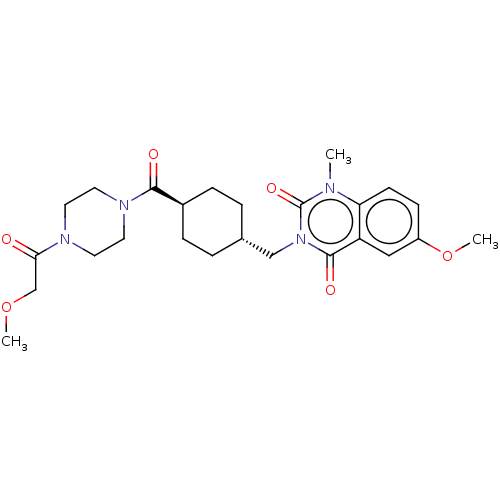
| ||