| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cytochrome P450 2D6 |
|---|
| Ligand | BDBM50192332 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1613494 (CHEMBL3855294) |
|---|
| IC50 | >10000±n/a nM |
|---|
| Citation |  Micheli, F; Bacchi, A; Braggio, S; Castelletti, L; Cavallini, P; Cavanni, P; Cremonesi, S; Dal Cin, M; Feriani, A; Gehanne, S; Kajbaf, M; Marchió, L; Nola, S; Oliosi, B; Pellacani, A; Perdoną, E; Sava, A; Semeraro, T; Tarsi, L; Tomelleri, S; Wong, A; Visentini, F; Zonzini, L; Heidbreder, C 1,2,4-Triazolyl 5-Azaspiro[2.4]heptanes: Lead Identification and Early Lead Optimization of a New Series of Potent and Selective Dopamine D3 Receptor Antagonists. J Med Chem59:8549-76 (2016) [PubMed] Article Micheli, F; Bacchi, A; Braggio, S; Castelletti, L; Cavallini, P; Cavanni, P; Cremonesi, S; Dal Cin, M; Feriani, A; Gehanne, S; Kajbaf, M; Marchió, L; Nola, S; Oliosi, B; Pellacani, A; Perdoną, E; Sava, A; Semeraro, T; Tarsi, L; Tomelleri, S; Wong, A; Visentini, F; Zonzini, L; Heidbreder, C 1,2,4-Triazolyl 5-Azaspiro[2.4]heptanes: Lead Identification and Early Lead Optimization of a New Series of Potent and Selective Dopamine D3 Receptor Antagonists. J Med Chem59:8549-76 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cytochrome P450 2D6 |
|---|
| Name: | Cytochrome P450 2D6 |
|---|
| Synonyms: | CP2D6_HUMAN | CYP2D6 | CYP2DL1 | CYPIID6 | Cytochrome P450 2D6 (CYP2D6) | Debrisoquine 4-hydroxylase | P450-DB1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 55774.82 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P10635 |
|---|
| Residue: | 497 |
|---|
| Sequence: | MGLEALVPLAVIVAIFLLLVDLMHRRQRWAARYPPGPLPLPGLGNLLHVDFQNTPYCFDQ
LRRRFGDVFSLQLAWTPVVVLNGLAAVREALVTHGEDTADRPPVPITQILGFGPRSQGVF
LARYGPAWREQRRFSVSTLRNLGLGKKSLEQWVTEEAACLCAAFANHSGRPFRPNGLLDK
AVSNVIASLTCGRRFEYDDPRFLRLLDLAQEGLKEESGFLREVLNAVPVLLHIPALAGKV
LRFQKAFLTQLDELLTEHRMTWDPAQPPRDLTEAFLAEMEKAKGNPESSFNDENLRIVVA
DLFSAGMVTTSTTLAWGLLLMILHPDVQRRVQQEIDDVIGQVRRPEMGDQAHMPYTTAVI
HEVQRFGDIVPLGVTHMTSRDIEVQGFRIPKGTTLITNLSSVLKDEAVWEKPFRFHPEHF
LDAQGHFVKPEAFLPFSAGRRACLGEPLARMELFLFFTSLLQHFSFSVPTGQPRPSHHGV
FAFLVSPSPYELCAVPR
|
|
|
|---|
| BDBM50192332 |
|---|
| n/a |
|---|
| Name | BDBM50192332 |
|---|
| Synonyms: | CHEMBL3948167 | US10239870, Example 70 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H31F3N4OS |
|---|
| Mol. Mass. | 480.589 |
|---|
| SMILES | Cn1c(SCCCN2CC[C@]3(C[C@@H]3c3ccc(cc3)C(F)(F)F)C2)nnc1C1CCOCC1 |r| |
|---|
| Structure | 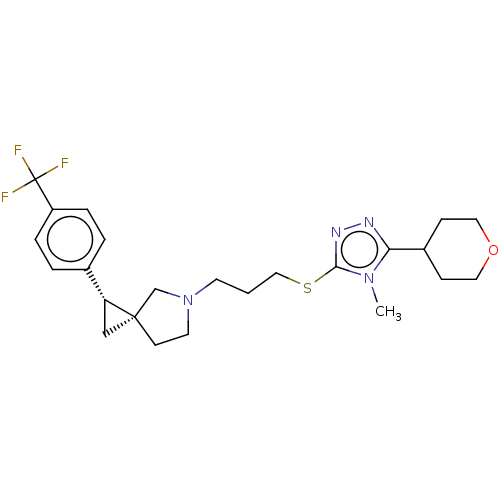
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Micheli, F; Bacchi, A; Braggio, S; Castelletti, L; Cavallini, P; Cavanni, P; Cremonesi, S; Dal Cin, M; Feriani, A; Gehanne, S; Kajbaf, M; Marchió, L; Nola, S; Oliosi, B; Pellacani, A; Perdoną, E; Sava, A; Semeraro, T; Tarsi, L; Tomelleri, S; Wong, A; Visentini, F; Zonzini, L; Heidbreder, C 1,2,4-Triazolyl 5-Azaspiro[2.4]heptanes: Lead Identification and Early Lead Optimization of a New Series of Potent and Selective Dopamine D3 Receptor Antagonists. J Med Chem59:8549-76 (2016) [PubMed] Article
Micheli, F; Bacchi, A; Braggio, S; Castelletti, L; Cavallini, P; Cavanni, P; Cremonesi, S; Dal Cin, M; Feriani, A; Gehanne, S; Kajbaf, M; Marchió, L; Nola, S; Oliosi, B; Pellacani, A; Perdoną, E; Sava, A; Semeraro, T; Tarsi, L; Tomelleri, S; Wong, A; Visentini, F; Zonzini, L; Heidbreder, C 1,2,4-Triazolyl 5-Azaspiro[2.4]heptanes: Lead Identification and Early Lead Optimization of a New Series of Potent and Selective Dopamine D3 Receptor Antagonists. J Med Chem59:8549-76 (2016) [PubMed] Article