| Reaction Details |
|---|
 | Report a problem with these data |
| Target | [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 1, mitochondrial |
|---|
| Ligand | BDBM227593 |
|---|
| Substrate/Competitor | PDC core E2/E3BP |
|---|
| Meas. Tech. | PDK Inhibition Assay |
|---|
| pH | 7.5±0 |
|---|
| Temperature | 298.15±0 K |
|---|
| IC50 | 2.14e+3± 3.3e+2 nM |
|---|
| Citation |  Tso, SC; Qi, X; Gui, WJ; Wu, CY; Chuang, JL; Wernstedt-Asterholm, I; Morlock, LK; Owens, KR; Scherer, PE; Williams, NS; Tambar, UK; Wynn, RM; Chuang, DT Structure-guided development of specific pyruvate dehydrogenase kinase inhibitors targeting the ATP-binding pocket. J Biol Chem289:4432-43 (2014) [PubMed] Article Tso, SC; Qi, X; Gui, WJ; Wu, CY; Chuang, JL; Wernstedt-Asterholm, I; Morlock, LK; Owens, KR; Scherer, PE; Williams, NS; Tambar, UK; Wynn, RM; Chuang, DT Structure-guided development of specific pyruvate dehydrogenase kinase inhibitors targeting the ATP-binding pocket. J Biol Chem289:4432-43 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 1, mitochondrial |
|---|
| Name: | [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 1, mitochondrial |
|---|
| Synonyms: | PDHK1 | PDK1 | PDK1_HUMAN | Pyruvate dehydrogenase kinase | Pyruvate dehydrogenase kinase 1 (PDK1) | Pyruvate dehydrogenase kinase isoform 1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 49255.23 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q15118 |
|---|
| Residue: | 436 |
|---|
| Sequence: | MRLARLLRGAALAGPGPGLRAAGFSRSFSSDSGSSPASERGVPGQVDFYARFSPSPLSMK
QFLDFGSVNACEKTSFMFLRQELPVRLANIMKEISLLPDNLLRTPSVQLVQSWYIQSLQE
LLDFKDKSAEDAKAIYDFTDTVIRIRNRHNDVIPTMAQGVIEYKESFGVDPVTSQNVQYF
LDRFYMSRISIRMLLNQHSLLFGGKGKGSPSHRKHIGSINPNCNVLEVIKDGYENARRLC
DLYYINSPELELEELNAKSPGQPIQVVYVPSHLYHMVFELFKNAMRATMEHHANRGVYPP
IQVHVTLGNEDLTVKMSDRGGGVPLRKIDRLFNYMYSTAPRPRVETSRAVPLAGFGYGLP
ISRLYAQYFQGDLKLYSLEGYGTDAVIYIKALSTDSIERLPVYNKAAWKHYNTNHEADDW
CVPSREPKDMTTFRSA
|
|
|
|---|
| BDBM227593 |
|---|
| Name | BDBM50003652 |
|---|
| Synonyms: | CHEMBL466897 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H21ClN4S |
|---|
| Mol. Mass. | 336.883 |
|---|
| SMILES | CCCCNC(=S)NCCNc1ccnc2cc(Cl)ccc12 |
|---|
| Structure | 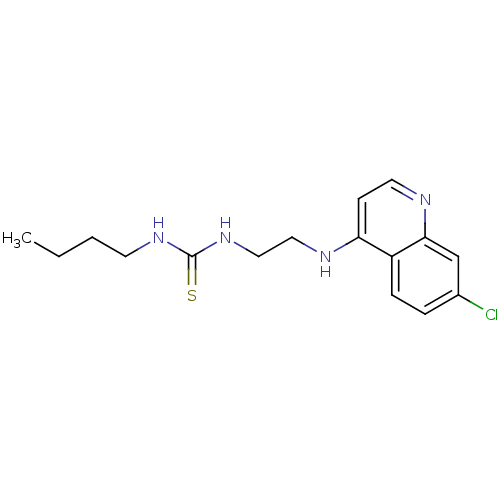
|
|---|
| PDC core E2/E3BP |
|---|
| Name: | PDC core E2/E3BP |
|---|
| Synonyms: | n/a |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | n/a |
|---|
| Components: | This complex has 2 components. |
|---|
| Component 1 |
| Name: | Pyruvate dehydrogenase complex component E2 |
|---|
| Synonyms: | DLAT | DLTA | ODP2_HUMAN |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 69001.42 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P10515 |
|---|
| Residue: | 647 |
|---|
| Sequence: | MWRVCARRAQNVAPWAGLEARWTALQEVPGTPRVTSRSGPAPARRNSVTTGYGGVRALCG
WTPSSGATPRNRLLLQLLGSPGRRYYSLPPHQKVPLPSLSPTMQAGTIARWEKKEGDKIN
EGDLIAEVETDKATVGFESLEECYMAKILVAEGTRDVPIGAIICITVGKPEDIEAFKNYT
LDSSAAPTPQAAPAPTPAATASPPTPSAQAPGSSYPPHMQVLLPALSPTMTMGTVQRWEK
KVGEKLSEGDLLAEIETDKATIGFEVQEEGYLAKILVPEGTRDVPLGTPLCIIVEKEADI
SAFADYRPTEVTDLKPQVPPPTPPPVAAVPPTPQPLAPTPSAPCPATPAGPKGRVFVSPL
AKKLAVEKGIDLTQVKGTGPDGRITKKDIDSFVPSKVAPAPAAVVPPTGPGMAPVPTGVF
TDIPISNIRRVIAQRLMQSKQTIPHYYLSIDVNMGEVLLVRKELNKILEGRSKISVNDFI
IKASALACLKVPEANSSWMDTVIRQNHVVDVSVAVSTPAGLITPIVFNAHIKGVETIAND
VVSLATKAREGKLQPHEFQGGTFTISNLGMFGIKNFSAIINPPQACILAIGASEDKLVPA
DNEKGFDVASMMSVTLSCDHRVVDGAVGAQWLAEFRKYLEKPITMLL
|
|
|
|---|
| Component 2 |
| Name: | Pyruvate dehydrogenase complex component E3-binding protein |
|---|
| Synonyms: | ODPX_HUMAN | PDHX | PDX1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 54129.22 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O00330 |
|---|
| Residue: | 501 |
|---|
| Sequence: | MAASWRLGCDPRLLRYLVGFPGRRSVGLVKGALGWSVSRGANWRWFHSTQWLRGDPIKIL
MPSLSPTMEEGNIVKWLKKEGEAVSAGDALCEIETDKAVVTLDASDDGILAKIVVEEGSK
NIRLGSLIGLIVEEGEDWKHVEIPKDVGPPPPVSKPSEPRPSPEPQISIPVKKEHIPGTL
RFRLSPAARNILEKHSLDASQGTATGPRGIFTKEDALKLVQLKQTGKITESRPTPAPTAT
PTAPSPLQATAGPSYPRPVIPPVSTPGQPNAVGTFTEIPASNIRRVIAKRLTESKSTVPH
AYATADCDLGAVLKVRQDLVKDDIKVSVNDFIIKAAAVTLKQMPDVNVSWDGEGPKQLPF
IDISVAVATDKGLLTPIIKDAAAKGIQEIADSVKALSKKARDGKLLPEEYQGGSFSISNL
GMFGIDEFTAVINPPQACILAVGRFRPVLKLTEDEEGNAKLQQRQLITVTMSSDSRVVDD
ELATRFLKSFKANLENPIRLA
|
|
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Tso, SC; Qi, X; Gui, WJ; Wu, CY; Chuang, JL; Wernstedt-Asterholm, I; Morlock, LK; Owens, KR; Scherer, PE; Williams, NS; Tambar, UK; Wynn, RM; Chuang, DT Structure-guided development of specific pyruvate dehydrogenase kinase inhibitors targeting the ATP-binding pocket. J Biol Chem289:4432-43 (2014) [PubMed] Article
Tso, SC; Qi, X; Gui, WJ; Wu, CY; Chuang, JL; Wernstedt-Asterholm, I; Morlock, LK; Owens, KR; Scherer, PE; Williams, NS; Tambar, UK; Wynn, RM; Chuang, DT Structure-guided development of specific pyruvate dehydrogenase kinase inhibitors targeting the ATP-binding pocket. J Biol Chem289:4432-43 (2014) [PubMed] Article