| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Replicase polyprotein 1ab [3264-3569] |
|---|
| Ligand | BDBM420295 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Determination of inhibition of SARS-CoV-2 Mproby 11r, 13a,and 13b |
|---|
| IC50 | 670±180 nM |
|---|
| Citation |  Zhang, L; Lin, D; Sun, X; Curth, U; Drosten, C; Sauerhering, L; Becker, S; Rox, K; Hilgenfeld, R Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved ?-ketoamide inhibitors. Science368:409-412 (2020) [PubMed] Article Zhang, L; Lin, D; Sun, X; Curth, U; Drosten, C; Sauerhering, L; Becker, S; Rox, K; Hilgenfeld, R Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved ?-ketoamide inhibitors. Science368:409-412 (2020) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Replicase polyprotein 1ab [3264-3569] |
|---|
| Name: | Replicase polyprotein 1ab [3264-3569] |
|---|
| Synonyms: | R1AB_SARS2 | Replicase polyprotein 1ab | rep |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | 33795.80 |
|---|
| Organism: | Severe acute respiratory syndrome coronavirus 2 |
|---|
| Description: | P0DTD1[3264-3569] |
|---|
| Residue: | 306 |
|---|
| Sequence: | SGFRKMAFPSGKVEGCMVQVTCGTTTLNGLWLDDVVYCPRHVICTSEDMLNPNYEDLLIR
KSNHNFLVQAGNVQLRVIGHSMQNCVLKLKVDTANPKTPKYKFVRIQPGQTFSVLACYNG
SPSGVYQCAMRPNFTIKGSFLNGSCGSVGFNIDYDCVSFCYMHHMELPTGVHAGTDLEGN
FYGPFVDRQTAQAAGTDTTITVNVLAWLYAAVINGDRWFLNRFTTTLNDFNLVAMKYNYE
PLTQDHVDILGPLSAQTGIAVLDMCASLKELLQNGMNGRTILGSALLEDEFTPFDVVRQC
SGVTFQ
|
|
|
|---|
| BDBM420295 |
|---|
| n/a |
|---|
| Name | BDBM420295 |
|---|
| Synonyms: | alpha-ketoamide inhibitor 13b | med.21724, Compound 67 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C31H39N5O7 |
|---|
| Mol. Mass. | 593.6707 |
|---|
| SMILES | CC(C)(C)OC(=O)Nc1cccn([C@@H](CC2CC2)C(=O)N[C@@H](C[C@@H]2CCNC2=O)C(=O)C(=O)NCc2ccccc2)c1=O |
|---|
| Structure | 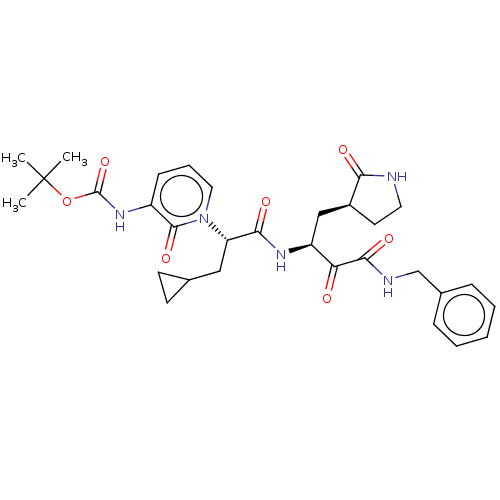
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Zhang, L; Lin, D; Sun, X; Curth, U; Drosten, C; Sauerhering, L; Becker, S; Rox, K; Hilgenfeld, R Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved ?-ketoamide inhibitors. Science368:409-412 (2020) [PubMed] Article
Zhang, L; Lin, D; Sun, X; Curth, U; Drosten, C; Sauerhering, L; Becker, S; Rox, K; Hilgenfeld, R Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved ?-ketoamide inhibitors. Science368:409-412 (2020) [PubMed] Article