| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Leucine-rich repeat serine/threonine-protein kinase 2 |
|---|
| Ligand | BDBM620948 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | LRRK2 Km ATP LanthaScreen Assay |
|---|
| IC50 | 0.625±n/a nM |
|---|
| Citation |  Acton, III, JJ; Chau, R; Fuller, PH; Gulati, A; Johnson, RE; Kattar, S; Keylor, MH; Li, D; Margrey, KA; Morriello, GJ; Yan, X 2-AMINOQUINAZOLINES AS LRRK2 INHIBITORS, PHARMACEUTICAL COMPOSITIONS, AND USES THEREOF US Patent US20230303540 Publication Date 9/28/2023 Acton, III, JJ; Chau, R; Fuller, PH; Gulati, A; Johnson, RE; Kattar, S; Keylor, MH; Li, D; Margrey, KA; Morriello, GJ; Yan, X 2-AMINOQUINAZOLINES AS LRRK2 INHIBITORS, PHARMACEUTICAL COMPOSITIONS, AND USES THEREOF US Patent US20230303540 Publication Date 9/28/2023 |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Leucine-rich repeat serine/threonine-protein kinase 2 |
|---|
| Name: | Leucine-rich repeat serine/threonine-protein kinase 2 |
|---|
| Synonyms: | Dardarin | LRRK2 | LRRK2_HUMAN | Leucine-Rich Repeat Kinase 2 Protein (LRRK2) | Leucine-rich repeat serine/threonine-protein kinase 2 | Leucine-rich repeat serine/threonine-protein kinase 2 (LRRK2) | PARK8 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 286113.20 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q5S007 |
|---|
| Residue: | 2527 |
|---|
| Sequence: | MASGSCQGCEEDEETLKKLIVRLNNVQEGKQIETLVQILEDLLVFTYSERASKLFQGKNI
HVPLLIVLDSYMRVASVQQVGWSLLCKLIEVCPGTMQSLMGPQDVGNDWEVLGVHQLILK
MLTVHNASVNLSVIGLKTLDLLLTSGKITLLILDEESDIFMLIFDAMHSFPANDEVQKLG
CKALHVLFERVSEEQLTEFVENKDYMILLSALTNFKDEEEIVLHVLHCLHSLAIPCNNVE
VLMSGNVRCYNIVVEAMKAFPMSERIQEVSCCLLHRLTLGNFFNILVLNEVHEFVVKAVQ
QYPENAALQISALSCLALLTETIFLNQDLEEKNENQENDDEGEEDKLFWLEACYKALTWH
RKNKHVQEAACWALNNLLMYQNSLHEKIGDEDGHFPAHREVMLSMLMHSSSKEVFQASAN
ALSTLLEQNVNFRKILLSKGIHLNVLELMQKHIHSPEVAESGCKMLNHLFEGSNTSLDIM
AAVVPKILTVMKRHETSLPVQLEALRAILHFIVPGMPEESREDTEFHHKLNMVKKQCFKN
DIHKLVLAALNRFIGNPGIQKCGLKVISSIVHFPDALEMLSLEGAMDSVLHTLQMYPDDQ
EIQCLGLSLIGYLITKKNVFIGTGHLLAKILVSSLYRFKDVAEIQTKGFQTILAILKLSA
SFSKLLVHHSFDLVIFHQMSSNIMEQKDQQFLNLCCKCFAKVAMDDYLKNVMLERACDQN
NSIMVECLLLLGADANQAKEGSSLICQVCEKESSPKLVELLLNSGSREQDVRKALTISIG
KGDSQIISLLLRRLALDVANNSICLGGFCIGKVEPSWLGPLFPDKTSNLRKQTNIASTLA
RMVIRYQMKSAVEEGTASGSDGNFSEDVLSKFDEWTFIPDSSMDSVFAQSDDLDSEGSEG
SFLVKKKSNSISVGEFYRDAVLQRCSPNLQRHSNSLGPIFDHEDLLKRKRKILSSDDSLR
SSKLQSHMRHSDSISSLASEREYITSLDLSANELRDIDALSQKCCISVHLEHLEKLELHQ
NALTSFPQQLCETLKSLTHLDLHSNKFTSFPSYLLKMSCIANLDVSRNDIGPSVVLDPTV
KCPTLKQFNLSYNQLSFVPENLTDVVEKLEQLILEGNKISGICSPLRLKELKILNLSKNH
ISSLSENFLEACPKVESFSARMNFLAAMPFLPPSMTILKLSQNKFSCIPEAILNLPHLRS
LDMSSNDIQYLPGPAHWKSLNLRELLFSHNQISILDLSEKAYLWSRVEKLHLSHNKLKEI
PPEIGCLENLTSLDVSYNLELRSFPNEMGKLSKIWDLPLDELHLNFDFKHIGCKAKDIIR
FLQQRLKKAVPYNRMKLMIVGNTGSGKTTLLQQLMKTKKSDLGMQSATVGIDVKDWPIQI
RDKRKRDLVLNVWDFAGREEFYSTHPHFMTQRALYLAVYDLSKGQAEVDAMKPWLFNIKA
RASSSPVILVGTHLDVSDEKQRKACMSKITKELLNKRGFPAIRDYHFVNATEESDALAKL
RKTIINESLNFKIRDQLVVGQLIPDCYVELEKIILSERKNVPIEFPVIDRKRLLQLVREN
QLQLDENELPHAVHFLNESGVLLHFQDPALQLSDLYFVEPKWLCKIMAQILTVKVEGCPK
HPKGIISRRDVEKFLSKKRKFPKNYMSQYFKLLEKFQIALPIGEEYLLVPSSLSDHRPVI
ELPHCENSEIIIRLYEMPYFPMGFWSRLINRLLEISPYMLSGRERALRPNRMYWRQGIYL
NWSPEAYCLVGSEVLDNHPESFLKITVPSCRKGCILLGQVVDHIDSLMEEWFPGLLEIDI
CGEGETLLKKWALYSFNDGEEHQKILLDDLMKKAEEGDLLVNPDQPRLTIPISQIAPDLI
LADLPRNIMLNNDELEFEQAPEFLLGDGSFGSVYRAAYEGEEVAVKIFNKHTSLRLLRQE
LVVLCHLHHPSLISLLAAGIRPRMLVMELASKGSLDRLLQQDKASLTRTLQHRIALHVAD
GLRYLHSAMIIYRDLKPHNVLLFTLYPNAAIIAKIADYGIAQYCCRMGIKTSEGTPGFRA
PEVARGNVIYNQQADVYSFGLLLYDILTTGGRIVEGLKFPNEFDELEIQGKLPDPVKEYG
CAPWPMVEKLIKQCLKENPQERPTSAQVFDILNSAELVCLTRRILLPKNVIVECMVATHH
NSRNASIWLGCGHTDRGQLSFLDLNTEGYTSEEVADSRILCLALVHLPVEKESWIVSGTQ
SGTLLVINTEDGKKRHTLEKMTDSVTCLYCNSFSKQSKQKNFLLVGTADGKLAIFEDKTV
KLKGAAPLKILNIGNVSTPLMCLSESTNSTERNVMWGGCGTKIFSFSNDFTIQKLIETRT
SQLFSYAAFSDSNIITVVVDTALYIAKQNSPVVEVWDKKTEKLCGLIDCVHFLREVMVKE
NKESKHKMSYSGRVKTLCLQKNTALWIGTGGGHILLLDLSTRRLIRVIYNFCNSVRVMMT
AQLGSLKNVMLVLGYNRKNTEGTQKQKEIQSCLTVWDINLPHEVQNLEKHIEVRKELAEK
MRRTSVE
|
|
|
|---|
| BDBM620948 |
|---|
| n/a |
|---|
| Name | BDBM620948 |
|---|
| Synonyms: | (1R,4S) or (1S,4R)-2-(2-((3- methyl-1-(2-(oxetan-3-yl)-2- azaspiro[3.3]heptan-6-yl)-1H- pyrazol-4-yl)amino)quinazolin- 7-yl)-2-azabicyclo[2.2.1]heptan- 3-one | US20230303540, Example Ex-8.5 | US20230303540, Example Ex-8.6 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H31N7O2 |
|---|
| Mol. Mass. | 485.5807 |
|---|
| SMILES | Cc1nn(cc1Nc1ncc2ccc(cc2n1)N1C2CCC(C2)C1=O)C1CC2(C1)CN(C2)C1COC1 |
|---|
| Structure | 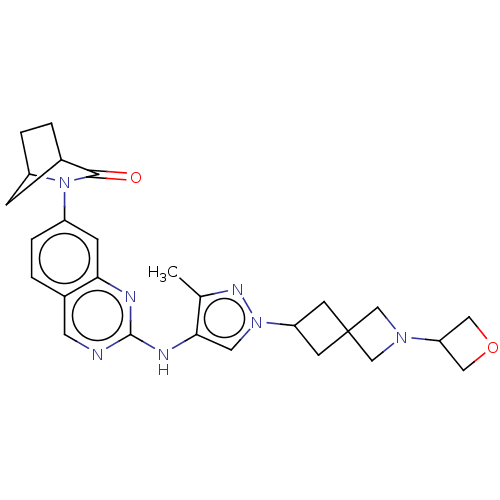
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Acton, III, JJ; Chau, R; Fuller, PH; Gulati, A; Johnson, RE; Kattar, S; Keylor, MH; Li, D; Margrey, KA; Morriello, GJ; Yan, X 2-AMINOQUINAZOLINES AS LRRK2 INHIBITORS, PHARMACEUTICAL COMPOSITIONS, AND USES THEREOF US Patent US20230303540 Publication Date 9/28/2023
Acton, III, JJ; Chau, R; Fuller, PH; Gulati, A; Johnson, RE; Kattar, S; Keylor, MH; Li, D; Margrey, KA; Morriello, GJ; Yan, X 2-AMINOQUINAZOLINES AS LRRK2 INHIBITORS, PHARMACEUTICAL COMPOSITIONS, AND USES THEREOF US Patent US20230303540 Publication Date 9/28/2023