| Reaction Details |
|---|
 | Report a problem with these data |
| Target | ADP-ribosylhydrolase ARH3 |
|---|
| Ligand | BDBM371049 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1793408 (CHEMBL4265327) |
|---|
| EC50 | >100000±n/a nM |
|---|
| Citation |  Waszkowycz, B; Smith, KM; McGonagle, AE; Jordan, AM; Acton, B; Fairweather, EE; Griffiths, LA; Hamilton, NM; Hamilton, NS; Hitchin, JR; Hutton, CP; James, DI; Jones, CD; Jones, S; Mould, DP; Small, HF; Stowell, AIJ; Tucker, JA; Waddell, ID; Ogilvie, DJ Cell-Active Small Molecule Inhibitors of the DNA-Damage Repair Enzyme Poly(ADP-ribose) Glycohydrolase (PARG): Discovery and Optimization of Orally Bioavailable Quinazolinedione Sulfonamides. J Med Chem61:10767-10792 (2018) [PubMed] Article Waszkowycz, B; Smith, KM; McGonagle, AE; Jordan, AM; Acton, B; Fairweather, EE; Griffiths, LA; Hamilton, NM; Hamilton, NS; Hitchin, JR; Hutton, CP; James, DI; Jones, CD; Jones, S; Mould, DP; Small, HF; Stowell, AIJ; Tucker, JA; Waddell, ID; Ogilvie, DJ Cell-Active Small Molecule Inhibitors of the DNA-Damage Repair Enzyme Poly(ADP-ribose) Glycohydrolase (PARG): Discovery and Optimization of Orally Bioavailable Quinazolinedione Sulfonamides. J Med Chem61:10767-10792 (2018) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| ADP-ribosylhydrolase ARH3 |
|---|
| Name: | ADP-ribosylhydrolase ARH3 |
|---|
| Synonyms: | ADP-ribose glycohydrolase ARH3 | ADP-ribosylhydrolase 3 | ADPRHL2 | ADPRS | ADPRS_HUMAN | ARH3 | O-acetyl-ADP-ribose deacetylase ARH3 | Poly(ADP-ribose) glycohydrolase ARH3 | [Protein ADP-ribosylarginine] hydrolase-like protein 2 | [Protein ADP-ribosylserine] hydrolase |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 38934.22 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9NX46 |
|---|
| Residue: | 363 |
|---|
| Sequence: | MAAAAMAAAAGGGAGAARSLSRFRGCLAGALLGDCVGSFYEAHDTVDLTSVLRHVQSLEP
DPGTPGSERTEALYYTDDTAMARALVQSLLAKEAFDEVDMAHRFAQEYKKDPDRGYGAGV
VTVFKKLLNPKCRDVFEPARAQFNGKGSYGNGGAMRVAGISLAYSSVQDVQKFARLSAQL
THASSLGYNGAILQALAVHLALQGESSSEHFLKQLLGHMEDLEGDAQSVLDARELGMEER
PYSSRLKKIGELLDQASVTREEVVSELGNGIAAFESVPTAIYCFLRCMEPDPEIPSAFNS
LQRTLIYSISLGGDTDTIATMAGAIAGAYYGMDQVPESWQQSCEGYEETDILAQSLHRVF
QKS
|
|
|
|---|
| BDBM371049 |
|---|
| n/a |
|---|
| Name | BDBM371049 |
|---|
| Synonyms: | US10239843, Example 156 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H24N4O4S2 |
|---|
| Mol. Mass. | 460.57 |
|---|
| SMILES | Cc1ncc(Cn2c(=O)n(CC3CC3)c3ccc(cc3c2=O)S(=O)(=O)NC2(C)CC2)s1 |
|---|
| Structure | 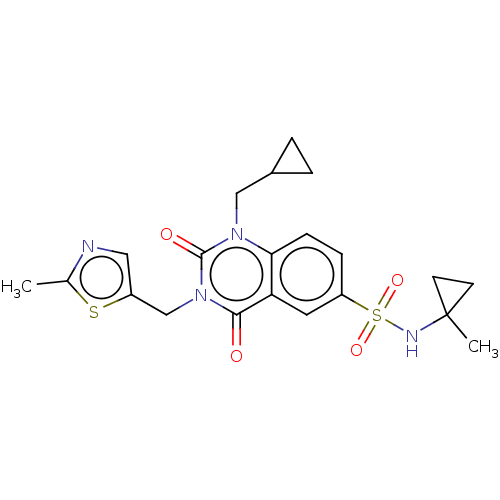
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Waszkowycz, B; Smith, KM; McGonagle, AE; Jordan, AM; Acton, B; Fairweather, EE; Griffiths, LA; Hamilton, NM; Hamilton, NS; Hitchin, JR; Hutton, CP; James, DI; Jones, CD; Jones, S; Mould, DP; Small, HF; Stowell, AIJ; Tucker, JA; Waddell, ID; Ogilvie, DJ Cell-Active Small Molecule Inhibitors of the DNA-Damage Repair Enzyme Poly(ADP-ribose) Glycohydrolase (PARG): Discovery and Optimization of Orally Bioavailable Quinazolinedione Sulfonamides. J Med Chem61:10767-10792 (2018) [PubMed] Article
Waszkowycz, B; Smith, KM; McGonagle, AE; Jordan, AM; Acton, B; Fairweather, EE; Griffiths, LA; Hamilton, NM; Hamilton, NS; Hitchin, JR; Hutton, CP; James, DI; Jones, CD; Jones, S; Mould, DP; Small, HF; Stowell, AIJ; Tucker, JA; Waddell, ID; Ogilvie, DJ Cell-Active Small Molecule Inhibitors of the DNA-Damage Repair Enzyme Poly(ADP-ribose) Glycohydrolase (PARG): Discovery and Optimization of Orally Bioavailable Quinazolinedione Sulfonamides. J Med Chem61:10767-10792 (2018) [PubMed] Article