| Reaction Details |
|---|
 | Report a problem with these data |
| Target | NAD-dependent protein deacetylase |
|---|
| Ligand | BDBM27507 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1822875 (CHEMBL4322639) |
|---|
| IC50 | 23100±n/a nM |
|---|
| Citation |  Monaldi, D; Rotili, D; Lancelot, J; Marek, M; Wössner, N; Lucidi, A; Tomaselli, D; Ramos-Morales, E; Romier, C; Pierce, RJ; Mai, A; Jung, M Structure-Reactivity Relationships on Substrates and Inhibitors of the Lysine Deacylase Sirtuin 2 from J Med Chem62:8733-8759 (2019) [PubMed] Article Monaldi, D; Rotili, D; Lancelot, J; Marek, M; Wössner, N; Lucidi, A; Tomaselli, D; Ramos-Morales, E; Romier, C; Pierce, RJ; Mai, A; Jung, M Structure-Reactivity Relationships on Substrates and Inhibitors of the Lysine Deacylase Sirtuin 2 from J Med Chem62:8733-8759 (2019) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| NAD-dependent protein deacetylase |
|---|
| Name: | NAD-dependent protein deacetylase |
|---|
| Synonyms: | 2.3.1.286 | NAD-dependent protein deacetylase | Sirt2 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 38395.78 |
|---|
| Organism: | Schistosoma mansoni |
|---|
| Description: | ChEMBL_119267 |
|---|
| Residue: | 337 |
|---|
| Sequence: | MSFDFLGIKKALFGDNTPRPELKSLNIEGVAQLIQDGQVNKIITMVGAGVSTAAGIPDFR
SPSSGIYDNLEDFNLPTPNAIFTIDYFRRDPRPFFEIARRLYRPEAKPTLAHCFIRLLHD
KGLLLRHYTQNVDSLERLSGLPEEKLVEAHGTFHTGHCIKCNKQHDFEFMLNEILAKRVP
QCLKCRNVVKPDVVLFGESMPKKFFKNLSSDLNNCDLLIIMGTSLTVLPFCAMIHRVGND
VPRLYINREYNDGSTESGLSSFIMRFMVAGFKQNYMKWGRSDNKRDIFWSGNADDGVVKI
SELLGWKDDLLRLKKETDSRLNEEFLAKKSQDKTNGQ
|
|
|
|---|
| BDBM27507 |
|---|
| n/a |
|---|
| Name | BDBM27507 |
|---|
| Synonyms: | 3-Pyridinecarboxamide | CHEMBL1140 | NAM | niacinamide | nicotinamide | pyridine-3-carboxamide |
|---|
| Type | n/a |
|---|
| Emp. Form. | C6H6N2O |
|---|
| Mol. Mass. | 122.1246 |
|---|
| SMILES | NC(=O)c1cccnc1 |
|---|
| Structure | 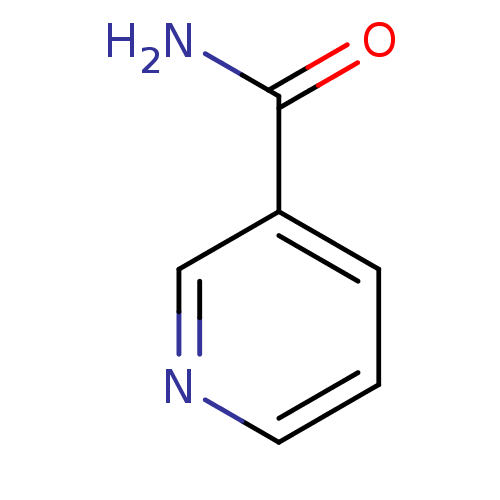
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Monaldi, D; Rotili, D; Lancelot, J; Marek, M; Wössner, N; Lucidi, A; Tomaselli, D; Ramos-Morales, E; Romier, C; Pierce, RJ; Mai, A; Jung, M Structure-Reactivity Relationships on Substrates and Inhibitors of the Lysine Deacylase Sirtuin 2 from J Med Chem62:8733-8759 (2019) [PubMed] Article
Monaldi, D; Rotili, D; Lancelot, J; Marek, M; Wössner, N; Lucidi, A; Tomaselli, D; Ramos-Morales, E; Romier, C; Pierce, RJ; Mai, A; Jung, M Structure-Reactivity Relationships on Substrates and Inhibitors of the Lysine Deacylase Sirtuin 2 from J Med Chem62:8733-8759 (2019) [PubMed] Article