

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Poly(ADP-ribose) glycohydrolase | ||
| Ligand | BDBM50541116 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1983746 (CHEMBL4617152) | ||
| IC50 | 29700±n/a nM | ||
| Citation |  Shibui, Y; Oyama, T; Okazawa, M; Yoshimori, A; Abe, H; Uchiumi, F; Tanuma, SI Structural insights into the active site of poly(ADP-ribose) glycohydrolase using docking modes of 6-hydroxy-3H-xanthen-3-one derivative inhibitors. Bioorg Med Chem28:0 (2020) [PubMed] Article Shibui, Y; Oyama, T; Okazawa, M; Yoshimori, A; Abe, H; Uchiumi, F; Tanuma, SI Structural insights into the active site of poly(ADP-ribose) glycohydrolase using docking modes of 6-hydroxy-3H-xanthen-3-one derivative inhibitors. Bioorg Med Chem28:0 (2020) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Poly(ADP-ribose) glycohydrolase | |||
| Name: | Poly(ADP-ribose) glycohydrolase | ||
| Synonyms: | PARG | PARG_HUMAN | Poly(ADP-ribose) glycohydrolase | poly(ADP-ribose) glycohydrolase (PARG) | ||
| Type: | Protein | ||
| Mol. Mass.: | 111107.13 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q86W56 | ||
| Residue: | 976 | ||
| Sequence: |
| ||
| BDBM50541116 | |||
| n/a | |||
| Name | BDBM50541116 | ||
| Synonyms: | CHEBI:87202 | Rose bengal | ||
| Type | Small organic molecule | ||
| Emp. Form. | C20H4Cl4I4O5 | ||
| Mol. Mass. | 973.673 | ||
| SMILES | OC(=O)c1c(Cl)c(Cl)c(Cl)c(Cl)c1-c1c2cc(I)c(O)c(I)c2oc2c(I)c(=O)c(I)cc12 |(2.68,-2.06,;2.67,-3.09,;3.74,-3.7,;1.34,-3.85,;1.34,-5.39,;2.4,-6.01,;0,-6.16,;0,-7.39,;-1.33,-5.39,;-2.4,-6.01,;-1.33,-3.85,;-2.4,-3.23,;0,-3.08,;,-1.54,;-1.33,-.77,;-2.67,-1.54,;-4,-.77,;-5.07,-1.39,;-4,.77,;-5.07,1.39,;-2.67,1.54,;-2.67,2.77,;-1.33,.77,;,1.54,;1.31,.77,;2.67,1.54,;2.67,2.77,;4,.77,;5.07,1.39,;4,-.77,;5.07,-1.39,;2.67,-1.54,;1.31,-.77,)| | ||
| Structure | 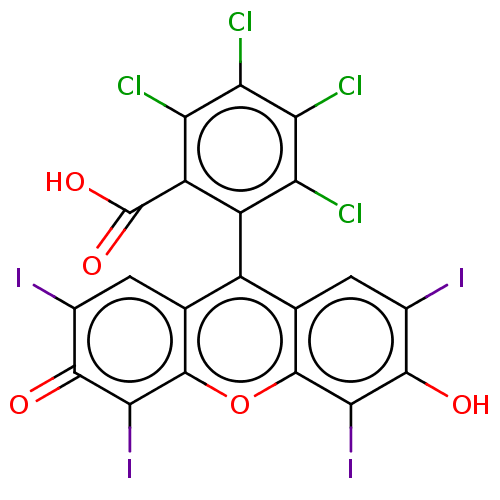
| ||