

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Bifunctional epoxide hydrolase 2 | ||
| Ligand | BDBM50223377 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_446700 (CHEMBL896996) | ||
| IC50 | 1.6±n/a nM | ||
| Citation |  Kim, IH; Tsai, HJ; Nishi, K; Kasagami, T; Morisseau, C; Hammock, BD 1,3-disubstituted ureas functionalized with ether groups are potent inhibitors of the soluble epoxide hydrolase with improved pharmacokinetic properties. J Med Chem50:5217-26 (2007) [PubMed] Article Kim, IH; Tsai, HJ; Nishi, K; Kasagami, T; Morisseau, C; Hammock, BD 1,3-disubstituted ureas functionalized with ether groups are potent inhibitors of the soluble epoxide hydrolase with improved pharmacokinetic properties. J Med Chem50:5217-26 (2007) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Bifunctional epoxide hydrolase 2 | |||
| Name: | Bifunctional epoxide hydrolase 2 | ||
| Synonyms: | Cytosolic epoxide hydrolase 2 | EBifunctional epoxide hydrolase 2 | EPHX2 | Epoxide hydratase | HYES_HUMAN | Lipid-phosphate phosphatase | Soluble epoxide hydrolase (sEH) | epoxide hydrolase 2, cytoplasmic | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 62613.07 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P34913 | ||
| Residue: | 555 | ||
| Sequence: |
| ||
| BDBM50223377 | |||
| n/a | |||
| Name | BDBM50223377 | ||
| Synonyms: | 1-adamantan-1-yl-3-{4-[2-(2-ethoxyethoxy)ethoxy]cyclohexyl}urea | CHEMBL244068 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H40N2O4 | ||
| Mol. Mass. | 408.5747 | ||
| SMILES | CCOCCOCCOC1CCC(CC1)NC(=O)NC12CC3CC(CC(C3)C1)C2 |TLB:18:19:22.21.26:24,THB:20:21:24:28.19.27,20:19:22.21.26:24,27:19:22:26.25.24,27:25:22:28.20.19,18:19:22:26.25.24,(31.61,-36.54,;30.89,-37.9,;29.35,-37.96,;28.63,-39.32,;27.09,-39.37,;26.37,-40.73,;24.83,-40.79,;24.11,-42.15,;22.57,-42.21,;21.75,-40.9,;20.21,-40.95,;19.4,-39.66,;20.12,-38.3,;21.66,-38.23,;22.47,-39.54,;19.3,-37,;17.76,-37.06,;16.94,-35.76,;17.04,-38.43,;15.51,-38.49,;14.49,-39.76,;13.08,-39.2,;11.59,-39.62,;12.78,-38.35,;12.77,-36.86,;14.12,-36.38,;13.08,-37.61,;15.52,-36.96,;14.11,-38.83,)| | ||
| Structure | 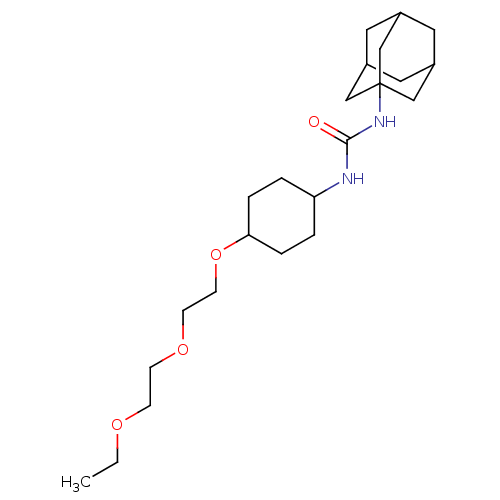
| ||