| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Fatty-acid amide hydrolase 1 |
|---|
| Ligand | BDBM50402685 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_886873 (CHEMBL2211371) |
|---|
| IC50 | 5.2±n/a nM |
|---|
| Citation |  Tichenor, MS; Keith, JM; Jones, WM; Pierce, JM; Merit, J; Hawryluk, N; Seierstad, M; Palmer, JA; Webb, M; Karbarz, MJ; Wilson, SJ; Wennerholm, ML; Woestenborghs, F; Beerens, D; Luo, L; Brown, SM; Boeck, MD; Chaplan, SR; Breitenbucher, JG Heteroaryl urea inhibitors of fatty acid amide hydrolase: structure-mutagenicity relationships for arylamine metabolites. Bioorg Med Chem Lett22:7357-62 (2012) [PubMed] Article Tichenor, MS; Keith, JM; Jones, WM; Pierce, JM; Merit, J; Hawryluk, N; Seierstad, M; Palmer, JA; Webb, M; Karbarz, MJ; Wilson, SJ; Wennerholm, ML; Woestenborghs, F; Beerens, D; Luo, L; Brown, SM; Boeck, MD; Chaplan, SR; Breitenbucher, JG Heteroaryl urea inhibitors of fatty acid amide hydrolase: structure-mutagenicity relationships for arylamine metabolites. Bioorg Med Chem Lett22:7357-62 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Fatty-acid amide hydrolase 1 |
|---|
| Name: | Fatty-acid amide hydrolase 1 |
|---|
| Synonyms: | Anandamide amidohydrolase | Anandamide amidohydrolase 1 | FAAH | FAAH1 | FAAH1_HUMAN | Fatty Acid Amide Hydrolase (FAAH) | Fatty-acid amide hydrolase (FAAH) | Fatty-acid amide hydrolase 1 | Oleamide hydrolase 1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 63071.19 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O00519 |
|---|
| Residue: | 579 |
|---|
| Sequence: | MVQYELWAALPGASGVALACCFVAAAVALRWSGRRTARGAVVRARQRQRAGLENMDRAAQ
RFRLQNPDLDSEALLALPLPQLVQKLHSRELAPEAVLFTYVGKAWEVNKGTNCVTSYLAD
CETQLSQAPRQGLLYGVPVSLKECFTYKGQDSTLGLSLNEGVPAECDSVVVHVLKLQGAV
PFVHTNVPQSMFSYDCSNPLFGQTVNPWKSSKSPGGSSGGEGALIGSGGSPLGLGTDIGG
SIRFPSSFCGICGLKPTGNRLSKSGLKGCVYGQEAVRLSVGPMARDVESLALCLRALLCE
DMFRLDPTVPPLPFREEVYTSSQPLRVGYYETDNYTMPSPAMRRAVLETKQSLEAAGHTL
VPFLPSNIPHALETLSTGGLFSDGGHTFLQNFKGDFVDPCLGDLVSILKLPQWLKGLLAF
LVKPLLPRLSAFLSNMKSRSAGKLWELQHEIEVYRKTVIAQWRALDLDVVLTPMLAPALD
LNAPGRATGAVSYTMLYNCLDFPAGVVPVTTVTAEDEAQMEHYRGYFGDIWDKMLQKGMK
KSVGLPVAVQCVALPWQEELCLRFMREVERLMTPEKQSS
|
|
|
|---|
| BDBM50402685 |
|---|
| n/a |
|---|
| Name | BDBM50402685 |
|---|
| Synonyms: | CHEMBL2207365 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H20Cl2N4O3 |
|---|
| Mol. Mass. | 447.314 |
|---|
| SMILES | Clc1conc1NC(=O)N1CCN(Cc2cccc(Oc3ccc(Cl)cc3)c2)CC1 |
|---|
| Structure | 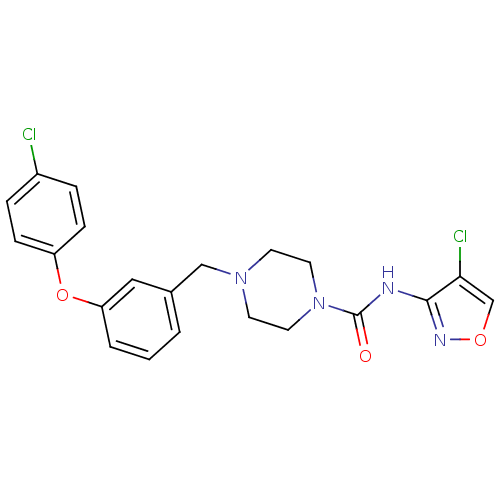
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Tichenor, MS; Keith, JM; Jones, WM; Pierce, JM; Merit, J; Hawryluk, N; Seierstad, M; Palmer, JA; Webb, M; Karbarz, MJ; Wilson, SJ; Wennerholm, ML; Woestenborghs, F; Beerens, D; Luo, L; Brown, SM; Boeck, MD; Chaplan, SR; Breitenbucher, JG Heteroaryl urea inhibitors of fatty acid amide hydrolase: structure-mutagenicity relationships for arylamine metabolites. Bioorg Med Chem Lett22:7357-62 (2012) [PubMed] Article
Tichenor, MS; Keith, JM; Jones, WM; Pierce, JM; Merit, J; Hawryluk, N; Seierstad, M; Palmer, JA; Webb, M; Karbarz, MJ; Wilson, SJ; Wennerholm, ML; Woestenborghs, F; Beerens, D; Luo, L; Brown, SM; Boeck, MD; Chaplan, SR; Breitenbucher, JG Heteroaryl urea inhibitors of fatty acid amide hydrolase: structure-mutagenicity relationships for arylamine metabolites. Bioorg Med Chem Lett22:7357-62 (2012) [PubMed] Article