| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Prostaglandin G/H synthase 2 |
|---|
| Ligand | BDBM50057943 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_157821 (CHEMBL858334) |
|---|
| IC50 | 3010000±n/a nM |
|---|
| Citation |  Desiraju, GR; Gopalakrishnan, B; Jetti, RK; Nagaraju, A; Raveendra, D; Sarma, JA; Sobhia, ME; Thilagavathi, R Computer-aided design of selective COX-2 inhibitors: comparative molecular field analysis, comparative molecular similarity indices analysis, and docking studies of some 1,2-diarylimidazole derivatives. J Med Chem45:4847-57 (2002) [PubMed] Desiraju, GR; Gopalakrishnan, B; Jetti, RK; Nagaraju, A; Raveendra, D; Sarma, JA; Sobhia, ME; Thilagavathi, R Computer-aided design of selective COX-2 inhibitors: comparative molecular field analysis, comparative molecular similarity indices analysis, and docking studies of some 1,2-diarylimidazole derivatives. J Med Chem45:4847-57 (2002) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Prostaglandin G/H synthase 2 |
|---|
| Name: | Prostaglandin G/H synthase 2 |
|---|
| Synonyms: | Cox-2 | Cox2 | Cyclooxygenase-2 | Cyclooxygenase-2 (COX-2) | Glucocorticoid-regulated inflammatory cyclooxygenase | Gripghs | Macrophage activation-associated marker protein P71/73 | PES-2 | PGH synthase 2 | PGH2_MOUSE | PGHS-2 | PHS II | Pghs-b | Prostaglandin G/H synthase (cyclooxygenase) | Prostaglandin H2 synthase 2 | Prostaglandin-endoperoxide synthase 2 | Ptgs2 | TIS10 protein | Tis10 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 69020.39 |
|---|
| Organism: | Mus musculus (Mouse) |
|---|
| Description: | Q05769 |
|---|
| Residue: | 604 |
|---|
| Sequence: | MLFRAVLLCAALGLSQAANPCCSNPCQNRGECMSTGFDQYKCDCTRTGFYGENCTTPEFL
TRIKLLLKPTPNTVHYILTHFKGVWNIVNNIPFLRSLIMKYVLTSRSYLIDSPPTYNVHY
GYKSWEAFSNLSYYTRALPPVADDCPTPMGVKGNKELPDSKEVLEKVLLRREFIPDPQGS
NMMFAFFAQHFTHQFFKTDHKRGPGFTRGLGHGVDLNHIYGETLDRQHKLRLFKDGKLKY
QVIGGEVYPPTVKDTQVEMIYPPHIPENLQFAVGQEVFGLVPGLMMYATIWLREHNRVCD
ILKQEHPEWGDEQLFQTSRLILIGETIKIVIEDYVQHLSGYHFKLKFDPELLFNQQFQYQ
NRIASEFNTLYHWHPLLPDTFNIEDQEYSFKQFLYNNSILLEHGLTQFVESFTRQIAGRV
AGGRNVPIAVQAVAKASIDQSREMKYQSLNEYRKRFSLKPYTSFEELTGEKEMAAELKAL
YSDIDVMELYPALLVEKPRPDAIFGETMVELGAPFSLKGLMGNPICSPQYWKPSTFGGEV
GFKIINTASIQSLICNNVKGCPFTSFNVQDPQPTKTATINASASHSRLDDINPTVLIKRR
STEL
|
|
|
|---|
| BDBM50057943 |
|---|
| n/a |
|---|
| Name | BDBM50057943 |
|---|
| Synonyms: | 4-[2-(3-Chloro-4-methyl-phenyl)-4-trifluoromethyl-imidazol-1-yl]-benzenesulfonamide | CHEMBL298476 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H13ClF3N3O2S |
|---|
| Mol. Mass. | 415.817 |
|---|
| SMILES | Cc1ccc(cc1Cl)-c1nc(cn1-c1ccc(cc1)S(N)(=O)=O)C(F)(F)F |
|---|
| Structure | 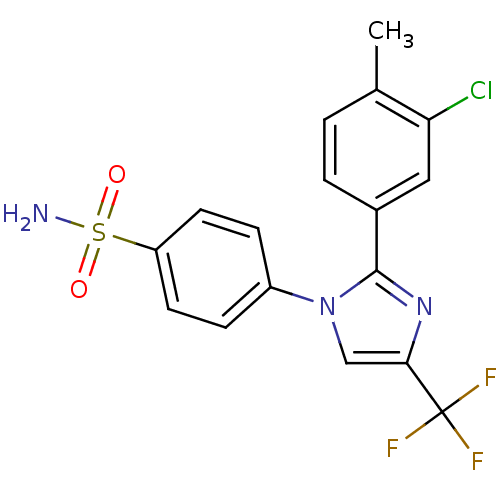
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Desiraju, GR; Gopalakrishnan, B; Jetti, RK; Nagaraju, A; Raveendra, D; Sarma, JA; Sobhia, ME; Thilagavathi, R Computer-aided design of selective COX-2 inhibitors: comparative molecular field analysis, comparative molecular similarity indices analysis, and docking studies of some 1,2-diarylimidazole derivatives. J Med Chem45:4847-57 (2002) [PubMed]
Desiraju, GR; Gopalakrishnan, B; Jetti, RK; Nagaraju, A; Raveendra, D; Sarma, JA; Sobhia, ME; Thilagavathi, R Computer-aided design of selective COX-2 inhibitors: comparative molecular field analysis, comparative molecular similarity indices analysis, and docking studies of some 1,2-diarylimidazole derivatives. J Med Chem45:4847-57 (2002) [PubMed]