| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Fatty-acid amide hydrolase 1 [33-574] |
|---|
| Ligand | BDBM108250 |
|---|
| Substrate/Competitor | n/a |
|---|
| IC50 | 60±10 nM |
|---|
| Citation |  Lambruschini, C; Veronesi, M; Romeo, E; Garau, G; Bandiera, T; Piomelli, D; Scarpelli, R; Dalvit, C Development of fragment-based n-FABS NMR screening applied to the membrane enzyme FAAH. Chembiochem14:1611-9 (2013) [PubMed] Article Lambruschini, C; Veronesi, M; Romeo, E; Garau, G; Bandiera, T; Piomelli, D; Scarpelli, R; Dalvit, C Development of fragment-based n-FABS NMR screening applied to the membrane enzyme FAAH. Chembiochem14:1611-9 (2013) [PubMed] Article |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| Fatty-acid amide hydrolase 1 [33-574] |
|---|
| Name: | Fatty-acid amide hydrolase 1 [33-574] |
|---|
| Synonyms: | FAAH1_RAT | Faah | Faah1 | Fatty-acid amide hydrolase (rFAAH∆TM) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 59459.86 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | 97-1722bp |
|---|
| Residue: | 542 |
|---|
| Sequence: | GRQKARGAATRARQKQRASLETMDKAVQRFRLQNPDLDSEALLTLPLLQLVQKLQSGELS
PEAVFFTYLGKAWEVNKGTNCVTSYLTDCETQLSQAPRQGLLYGVPVSLKECFSYKGHDS
TLGLSLNEGMPSESDCVVVQVLKLQGAVPFVHTNVPQSMLSFDCSNPLFGQTMNPWKSSK
SPGGSSGGEGALIGSGGSPLGLGTDIGGSIRFPSAFCGICGLKPTGNRLSKSGLKGCVYG
QTAVQLSLGPMARDVESLALCLKALLCEHLFTLDPTVPPLPFREEVYRSSRPLRVGYYET
DNYTMPSPAMRRALIETKQRLEAAGHTLIPFLPNNIPYALEVLSAGGLFSDGGRSFLQNF
KGDFVDPCLGDLILILRLPSWFKRLLSLLLKPLFPRLAAFLNSMRPRSAEKLWKLQHEIE
MYRQSVIAQWKAMNLDVLLTPMLGPALDLNTPGRATGAISYTVLYNCLDFPAGVVPVTTV
TAEDDAQMELYKGYFGDIWDIILKKAMKNSVGLPVAVQCVALPWQEELCLRFMREVEQLM
TP
|
|
|
|---|
| BDBM108250 |
|---|
| n/a |
|---|
| Name | BDBM108250 |
|---|
| Synonyms: | 3‐(3‐carbamoylphenyl)‐4‐methylphenyl N‐ cyclohexylcarbamate (Compound 1) | US9187413, 1d |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H24N2O3 |
|---|
| Mol. Mass. | 352.4269 |
|---|
| SMILES | Cc1ccc(OC(=O)NC2CCCCC2)cc1-c1cccc(c1)C(N)=O |
|---|
| Structure | 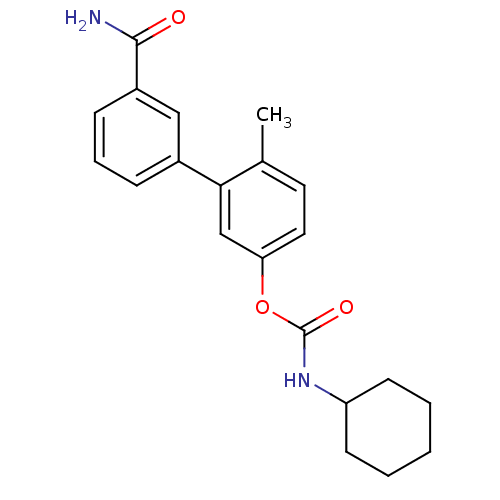
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lambruschini, C; Veronesi, M; Romeo, E; Garau, G; Bandiera, T; Piomelli, D; Scarpelli, R; Dalvit, C Development of fragment-based n-FABS NMR screening applied to the membrane enzyme FAAH. Chembiochem14:1611-9 (2013) [PubMed] Article
Lambruschini, C; Veronesi, M; Romeo, E; Garau, G; Bandiera, T; Piomelli, D; Scarpelli, R; Dalvit, C Development of fragment-based n-FABS NMR screening applied to the membrane enzyme FAAH. Chembiochem14:1611-9 (2013) [PubMed] Article