

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | E3 ubiquitin-protein ligase Mdm2 | ||
| Ligand | BDBM535783 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | Binding Assay (ELISA) | ||
| IC50 | 0.480±n/a nM | ||
| Citation |  Chessari, G; Lyons, JF Combination of isoindolinone derivatives with SGI-110 US Patent US11236047 Publication Date 2/1/2022 Chessari, G; Lyons, JF Combination of isoindolinone derivatives with SGI-110 US Patent US11236047 Publication Date 2/1/2022 | ||
| More Info.: | Get all data from this article, Assay Method | ||
| E3 ubiquitin-protein ligase Mdm2 | |||
| Name: | E3 ubiquitin-protein ligase Mdm2 | ||
| Synonyms: | Double minute 2 protein | Double minute 2 protein (HDM2) | E3 ubiquitin-protein ligase Mdm2 (p53-binding protein Mdm2) | Hdm2 | Human Double Minute 2 (HDM2) | MDM2 | MDM2-MDMX | MDM2_HUMAN | p53-Binding Protein MDM2 | p53-binding protein | ||
| Type: | Oncoprotein | ||
| Mol. Mass.: | 55196.54 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q00987 | ||
| Residue: | 491 | ||
| Sequence: |
| ||
| BDBM535783 | |||
| n/a | |||
| Name | BDBM535783 | ||
| Synonyms: | 2-{[(1R)-1-(4-chlorophenyl)-7-fluoro- 5-{1-hydroxy-1-[trans-4- hydroxycyclohexyl]propyl}-1-[(2R)-2- hydroxypropoxy]-3-oxo-2,3-dihydro- 1H-isoindol-2-yl]methyl}pyrimidine-5- carbonitrile | US11236047, Example 512 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C32H34ClFN4O5 | ||
| Mol. Mass. | 609.088 | ||
| SMILES | CCC(O)([C@H]1CC[C@H](O)CC1)c1cc2C(=O)N(Cc3ncc(cn3)C#N)[C@](OC[C@@H](C)O)(c2c(F)c1)c1ccc(Cl)cc1 |r,wU:4.3,26.28,29.32,wD:7.7,(-5.52,-2.86,;-3.98,-2.86,;-3.21,-1.53,;-2.44,-2.86,;-4.55,-.76,;-5.88,-1.53,;-7.21,-.76,;-7.21,.78,;-8.55,1.55,;-5.88,1.55,;-4.55,.78,;-1.88,-.76,;-.55,-1.53,;.79,-.76,;2.25,-1.23,;2.73,-2.7,;3.16,.01,;4.7,.01,;5.47,-1.32,;4.7,-2.65,;5.47,-3.99,;7.04,-4.04,;7.78,-2.65,;7.01,-1.32,;7.81,-5.37,;8.58,-6.7,;2.25,1.26,;1.48,2.59,;1.88,4.08,;.79,5.17,;-.7,4.77,;1.19,6.66,;.79,.78,;-.55,1.55,;-.55,3.09,;-1.88,.78,;3.59,2.03,;4.92,1.26,;6.25,2.03,;6.25,3.57,;7.59,4.34,;4.92,4.34,;3.59,3.57,)| | ||
| Structure | 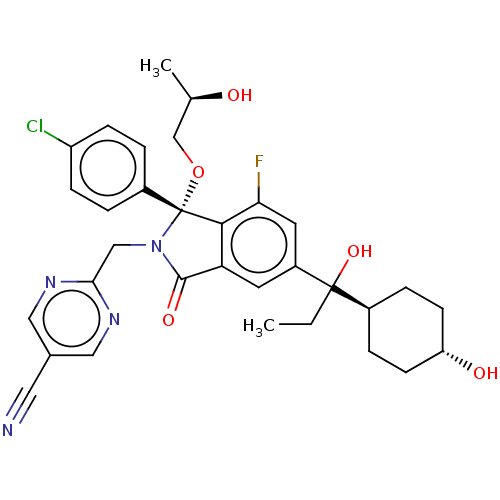
| ||