| Reaction Details |
|---|
 | Report a problem with these data |
| Target | MAP kinase-interacting serine/threonine-protein kinase 1 |
|---|
| Ligand | BDBM168250 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Mnk Biochemical Enzymatic Assay |
|---|
| Temperature | 298.15±n/a K |
|---|
| IC50 | <10.00±n/a nM |
|---|
| Comments | extracted |
|---|
| Citation |  Reich, SH; Sprengeler, PA; Webber, SE; Xiang, AX; Ernst, JT MNK inhibitors and methods related thereto US Patent US9669031 Publication Date 6/6/2017 Reich, SH; Sprengeler, PA; Webber, SE; Xiang, AX; Ernst, JT MNK inhibitors and methods related thereto US Patent US9669031 Publication Date 6/6/2017 |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| MAP kinase-interacting serine/threonine-protein kinase 1 |
|---|
| Name: | MAP kinase-interacting serine/threonine-protein kinase 1 |
|---|
| Synonyms: | MAP Kinase-Interacting Protein Kinase (MNK1) | MAP kinase signal-integrating kinase 1 | MAP kinase-interacting serine/threonine-protein kinase 1 (MnK1) | MAP kinase-interacting serine/threonine-protein kinase MNK1 | MAP-kinase interacting kinase 1 (MNK1) | MAPK signal-integrating kinase 1 | MKNK1 | MKNK1_HUMAN | MNK1 |
|---|
| Type: | Serine/threonine-protein kinase |
|---|
| Mol. Mass.: | 51342.85 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9BUB5 |
|---|
| Residue: | 465 |
|---|
| Sequence: | MVSSQKLEKPIEMGSSEPLPIADGDRRRKKKRRGRATDSLPGKFEDMYKLTSELLGEGAY
AKVQGAVSLQNGKEYAVKIIEKQAGHSRSRVFREVETLYQCQGNKNILELIEFFEDDTRF
YLVFEKLQGGSILAHIQKQKHFNEREASRVVRDVAAALDFLHTKDKVSLCHLGWSAMAPS
GLTAAPTSLGSSDPPTSASQVAGTTGIAHRDLKPENILCESPEKVSPVKICDFDLGSGMK
LNNSCTPITTPELTTPCGSAEYMAPEVVEVFTDQATFYDKRCDLWSLGVVLYIMLSGYPP
FVGHCGADCGWDRGEVCRVCQNKLFESIQEGKYEFPDKDWAHISSEAKDLISKLLVRDAK
QRLSAAQVLQHPWVQGQAPEKGLPTPQVLQRNSSTMDLTLFAAEAIALNRQLSQHEENEL
AEEPEALADGLCSMKLSPPCKSRLARRRALAQAGRGEDRSPPTAL
|
|
|
|---|
| BDBM168250 |
|---|
| n/a |
|---|
| Name | BDBM168250 |
|---|
| Synonyms: | US9669031, 78 N-(6-((8′-chloro-2-fluoro-1′,5′-dioxo-1′,5′-dihydro-2′H-spiro[cyclohexane-1,3′-imidazo[1,5-a]pyridin]-6′-yl)amino)pyrimidin-4-yl)cyclopropanecarboxamide (Cpd. No. 78) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H20ClFN6O3 |
|---|
| Mol. Mass. | 446.863 |
|---|
| SMILES | FC1CCCCC11NC(=O)c2c(Cl)cc(Nc3cc(NC(=O)C4CC4)ncn3)c(=O)n12 |
|---|
| Structure | 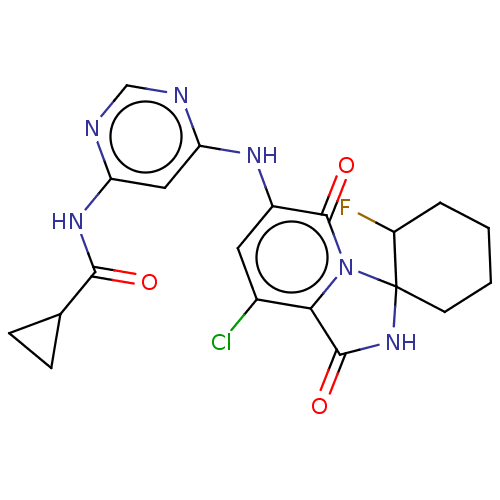
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Reich, SH; Sprengeler, PA; Webber, SE; Xiang, AX; Ernst, JT MNK inhibitors and methods related thereto US Patent US9669031 Publication Date 6/6/2017
Reich, SH; Sprengeler, PA; Webber, SE; Xiang, AX; Ernst, JT MNK inhibitors and methods related thereto US Patent US9669031 Publication Date 6/6/2017