| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Glutamate receptor ionotropic, NMDA 1 |
|---|
| Ligand | BDBM356628 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Protective Effects of Compounds on Primary Cerebellum Granule Cells of Rats |
|---|
| EC50 | 5200±n/a nM |
|---|
| Citation |  Wang, Y; Liu, Z; Yu, P; Sun, Y; Zhang, Z; Zhang, G; Shan, L; Yi, P; Larrick, J Amantadine nitrate compounds with neural protective effect, and preparation and medical use thereof US Patent US10214478 Publication Date 2/26/2019 Wang, Y; Liu, Z; Yu, P; Sun, Y; Zhang, Z; Zhang, G; Shan, L; Yi, P; Larrick, J Amantadine nitrate compounds with neural protective effect, and preparation and medical use thereof US Patent US10214478 Publication Date 2/26/2019 |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Glutamate receptor ionotropic, NMDA 1 |
|---|
| Name: | Glutamate receptor ionotropic, NMDA 1 |
|---|
| Synonyms: | Glutamate (NMDA) receptor subunit zeta 1 | Glutamate [NMDA] receptor subunit zeta-1 | Glutamate-NMDA-Channel | Glutamate-NMDA-MK801 | Glutamate-NMDA-Polyamine | Grin1 | NMDA | NMDZ1_RAT | Nmdar1 | phencyclidine |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 105533.40 |
|---|
| Organism: | RAT |
|---|
| Description: | P35439 |
|---|
| Residue: | 938 |
|---|
| Sequence: | MSTMHLLTFALLFSCSFARAACDPKIVNIGAVLSTRKHEQMFREAVNQANKRHGSWKIQL
NATSVTHKPNAIQMALSVCEDLISSQVYAILVSHPPTPNDHFTPTPVSYTAGFYRIPVLG
LTTRMSIYSDKSIHLSFLRTVPPYSHQSSVWFEMMRVYNWNHIILLVSDDHEGRAAQKRL
ETLLEERESKAEKVLQFDPGTKNVTALLMEARELEARVIILSASEDDAATVYRAAAMLNM
TGSGYVWLVGEREISGNALRYAPDGIIGLQLINGKNESAHISDAVGVVAQAVHELLEKEN
ITDPPRGCVGNTNIWKTGPLFKRVLMSSKYADGVTGRVEFNEDGDRKFANYSIMNLQNRK
LVQVGIYNGTHVIPNDRKIIWPGGETEKPRGYQMSTRLKIVTIHQEPFVYVKPTMSDGTC
KEEFTVNGDPVKKVICTGPNDTSPGSPRHTVPQCCYGFCIDLLIKLARTMNFTYEVHLVA
DGKFGTQERVNNSNKKEWNGMMGELLSGQADMIVAPLTINNERAQYIEFSKPFKYQGLTI
LVKKEIPRSTLDSFMQPFQSTLWLLVGLSVHVVAVMLYLLDRFSPFGRFKVNSEEEEEDA
LTLSSAMWFSWGVLLNSGIGEGAPRSFSARILGMVWAGFAMIIVASYTANLAAFLVLDRP
EERITGINDPRLRNPSDKFIYATVKQSSVDIYFRRQVELSTMYRHMEKHNYESAAEAIQA
VRDNKLHAFIWDSAVLEFEASQKCDLVTTGELFFRSGFGIGMRKDSPWKQNVSLSILKSH
ENGFMEDLDKTWVRYQECDSRSNAPATLTFENMAGVFMLVAGGIVAGIFLIFIEIAYKRH
KDARRKQMQLAFAAVNVWRKNLQDRKSGRAEPDPKKKATFRAITSTLASSFKRRRSSKDT
STGGGRGALQNQKDTVLPRRAIEREEGQLQLCSRHRES
|
|
|
|---|
| BDBM356628 |
|---|
| n/a |
|---|
| Name | BDBM356628 |
|---|
| Synonyms: | US10214478, Compound NM-009 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H23N3O6 |
|---|
| Mol. Mass. | 329.3489 |
|---|
| SMILES | NC12CC3CC(CCO[N+]([O-])=O)(C1)CC(CCO[N+]([O-])=O)(C3)C2 |TLB:12:1:21:5.4.13,4:5:22:21.3.2,4:3:22:5.12.13,6:5:21:22.1.2,THB:12:5:21:22.1.2| |
|---|
| Structure | 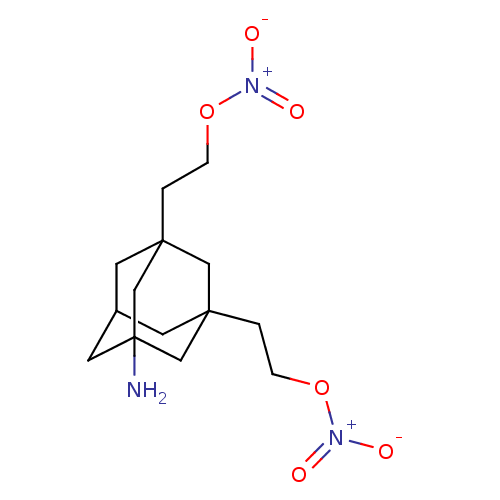
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wang, Y; Liu, Z; Yu, P; Sun, Y; Zhang, Z; Zhang, G; Shan, L; Yi, P; Larrick, J Amantadine nitrate compounds with neural protective effect, and preparation and medical use thereof US Patent US10214478 Publication Date 2/26/2019
Wang, Y; Liu, Z; Yu, P; Sun, Y; Zhang, Z; Zhang, G; Shan, L; Yi, P; Larrick, J Amantadine nitrate compounds with neural protective effect, and preparation and medical use thereof US Patent US10214478 Publication Date 2/26/2019