| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Poly(ADP-ribose) glycohydrolase |
|---|
| Ligand | BDBM371494 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | PARG Assay |
|---|
| IC50 | 29.0±n/a nM |
|---|
| Citation |  McGonagle, AE; Jordan, A; Waszkowycz, B; Hutton, C; Waddell, I; Hitchin, JR; Smith, KM; Hamilton, NM 2,4-dioxo-quinazoline-6-sulfonamide derivatives as inhibitors of PARG US Patent US10239843 Publication Date 3/26/2019 McGonagle, AE; Jordan, A; Waszkowycz, B; Hutton, C; Waddell, I; Hitchin, JR; Smith, KM; Hamilton, NM 2,4-dioxo-quinazoline-6-sulfonamide derivatives as inhibitors of PARG US Patent US10239843 Publication Date 3/26/2019 |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Poly(ADP-ribose) glycohydrolase |
|---|
| Name: | Poly(ADP-ribose) glycohydrolase |
|---|
| Synonyms: | PARG | PARG_HUMAN | Poly(ADP-ribose) glycohydrolase | poly(ADP-ribose) glycohydrolase (PARG) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 111107.13 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q86W56 |
|---|
| Residue: | 976 |
|---|
| Sequence: | MNAGPGCEPCTKRPRWGAATTSPAASDARSFPSRQRRVLDPKDAHVQFRVPPSSPACVPG
RAGQHRGSATSLVFKQKTITSWMDTKGIKTAESESLDSKENNNTRIESMMSSVQKDNFYQ
HNVEKLENVSQLSLDKSPTEKSTQYLNQHQTAAMCKWQNEGKHTEQLLESEPQTVTLVPE
QFSNANIDRSPQNDDHSDTDSEENRDNQQFLTTVKLANAKQTTEDEQAREAKSHQKCSKS
CDPGEDCASCQQDEIDVVPESPLSDVGSEDVGTGPKNDNKLTRQESCLGNSPPFEKESEP
ESPMDVDNSKNSCQDSEADEETSPGFDEQEDGSSSQTANKPSRFQARDADIEFRKRYSTK
GGEVRLHFQFEGGESRTGMNDLNAKLPGNISSLNVECRNSKQHGKKDSKITDHFMRLPKA
EDRRKEQWETKHQRTERKIPKYVPPHLSPDKKWLGTPIEEMRRMPRCGIRLPLLRPSANH
TVTIRVDLLRAGEVPKPFPTHYKDLWDNKHVKMPCSEQNLYPVEDENGERTAGSRWELIQ
TALLNKFTRPQNLKDAILKYNVAYSKKWDFTALIDFWDKVLEEAEAQHLYQSILPDMVKI
ALCLPNICTQPIPLLKQKMNHSITMSQEQIASLLANAFFCTFPRRNAKMKSEYSSYPDIN
FNRLFEGRSSRKPEKLKTLFCYFRRVTEKKPTGLVTFTRQSLEDFPEWERCEKPLTRLHV
TYEGTIEENGQGMLQVDFANRFVGGGVTSAGLVQEEIRFLINPELIISRLFTEVLDHNEC
LIITGTEQYSEYTGYAETYRWSRSHEDGSERDDWQRRCTEIVAIDALHFRRYLDQFVPEK
MRRELNKAYCGFLRPGVSSENLSAVATGNWGCGAFGGDARLKALIQILAAAAAERDVVYF
TFGDSELMRDIYSMHIFLTERKLTVGDVYKLLLRYYNEECRNCSTPGPDIKLYPFIYHAV
ESCAETADHSGQRTGT
|
|
|
|---|
| BDBM371494 |
|---|
| n/a |
|---|
| Name | BDBM371494 |
|---|
| Synonyms: | 1-[(2,5-Dimethylpyrazol-3-yl)methyl]-N-[1-(fluoromethyl)cyclopropyl]-3-[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]-2,4-dioxo-quinazoline-6-sulfonamide | US10239843, Example 603 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H24FN7O4S2 |
|---|
| Mol. Mass. | 533.599 |
|---|
| SMILES | Cc1cc(Cn2c3ccc(cc3c(=O)n(Cc3nnc(C)s3)c2=O)S(=O)(=O)NC2(CF)CC2)n(C)n1 |
|---|
| Structure | 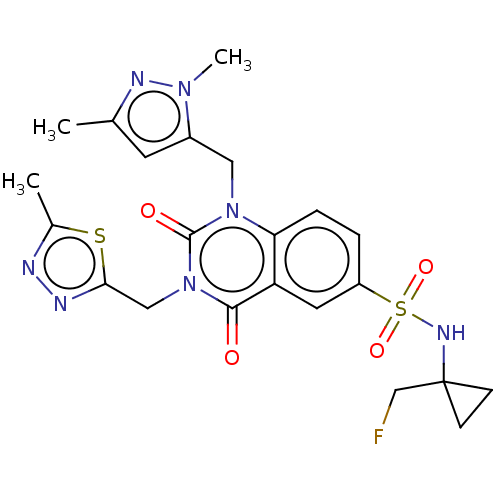
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 McGonagle, AE; Jordan, A; Waszkowycz, B; Hutton, C; Waddell, I; Hitchin, JR; Smith, KM; Hamilton, NM 2,4-dioxo-quinazoline-6-sulfonamide derivatives as inhibitors of PARG US Patent US10239843 Publication Date 3/26/2019
McGonagle, AE; Jordan, A; Waszkowycz, B; Hutton, C; Waddell, I; Hitchin, JR; Smith, KM; Hamilton, NM 2,4-dioxo-quinazoline-6-sulfonamide derivatives as inhibitors of PARG US Patent US10239843 Publication Date 3/26/2019