| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Purine nucleoside phosphorylase |
|---|
| Ligand | BDBM22108 |
|---|
| Substrate/Competitor | BDBM22104 |
|---|
| Meas. Tech. | PNP Inhibition Assay |
|---|
| pH | 7.7±n/a |
|---|
| Temperature | 295.15±n/a K |
|---|
| Ki | 1.8±0.3 nM |
|---|
| Km | 40000±n/a nM |
|---|
| Comments | Ki is the dissociation constant for the first step in the two-step binding characteristic of slow-onset tight-binding inhibition. |
|---|
| Citation |  Evans, GB; Furneaux, RH; Greatrex, B; Murkin, AS; Schramm, VL; Tyler, PC Azetidine based transition state analogue inhibitors of N-ribosyl hydrolases and phosphorylases. J Med Chem51:948-56 (2008) [PubMed] Article Evans, GB; Furneaux, RH; Greatrex, B; Murkin, AS; Schramm, VL; Tyler, PC Azetidine based transition state analogue inhibitors of N-ribosyl hydrolases and phosphorylases. J Med Chem51:948-56 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Purine nucleoside phosphorylase |
|---|
| Name: | Purine nucleoside phosphorylase |
|---|
| Synonyms: | Inosine phosphorylase | Inosine-guanosine phosphorylase | NP | PNP | PNPH_HUMAN | Purine nucleoside phosphorylase (PNPase) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 32119.53 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 289 |
|---|
| Sequence: | MENGYTYEDYKNTAEWLLSHTKHRPQVAIICGSGLGGLTDKLTQAQIFDYGEIPNFPRST
VPGHAGRLVFGFLNGRACVMMQGRFHMYEGYPLWKVTFPVRVFHLLGVDTLVVTNAAGGL
NPKFEVGDIMLIRDHINLPGFSGQNPLRGPNDERFGDRFPAMSDAYDRTMRQRALSTWKQ
MGEQRELQEGTYVMVAGPSFETVAECRVLQKLGADAVGMSTVPEVIVARHCGLRVFGFSL
ITNKVIMDYESLEKANHEEVLAAGKQAAQKLEQFVSILMASIPLPDKAS
|
|
|
|---|
| BDBM22108 |
|---|
| BDBM22104 |
|---|
| Name | BDBM22108 |
|---|
| Synonyms: | 7-{[2-(hydroxymethyl)azetidin-1-yl]methyl}-3H,4H,5H-pyrrolo[3,2-d]pyrimidin-4-one | Azetidine based compound, (+/-) 42 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C11H14N4O2 |
|---|
| Mol. Mass. | 234.2545 |
|---|
| SMILES | OCC1CCN1Cc1c[nH]c2c1nc[nH]c2=O |
|---|
| Structure | 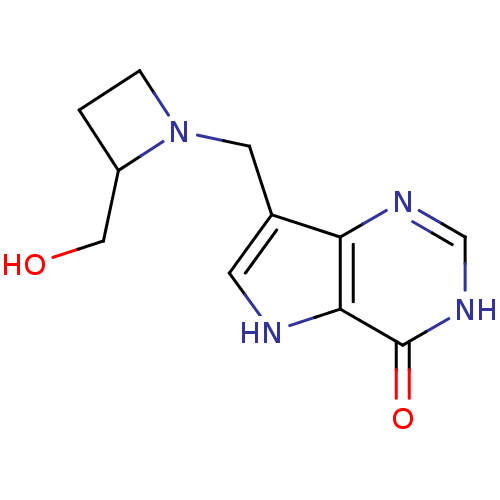
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Evans, GB; Furneaux, RH; Greatrex, B; Murkin, AS; Schramm, VL; Tyler, PC Azetidine based transition state analogue inhibitors of N-ribosyl hydrolases and phosphorylases. J Med Chem51:948-56 (2008) [PubMed] Article
Evans, GB; Furneaux, RH; Greatrex, B; Murkin, AS; Schramm, VL; Tyler, PC Azetidine based transition state analogue inhibitors of N-ribosyl hydrolases and phosphorylases. J Med Chem51:948-56 (2008) [PubMed] Article