| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 4 [653-1084,H976Y] |
|---|
| Ligand | BDBM19149 |
|---|
| Substrate/Competitor | BDBM25143 |
|---|
| Meas. Tech. | HDAC Activity Assay |
|---|
| Temperature | 298.15±n/a K |
|---|
| IC50 | 540±n/a nM |
|---|
| Comments | extracted |
|---|
| Citation |  Jones, P; Altamura, S; De Francesco, R; Paz, OG; Kinzel, O; Mesiti, G; Monteagudo, E; Pescatore, G; Rowley, M; Verdirame, M; Steinkühler, C A novel series of potent and selective ketone histone deacetylase inhibitors with antitumor activity in vivo. J Med Chem51:2350-3 (2008) [PubMed] Article Jones, P; Altamura, S; De Francesco, R; Paz, OG; Kinzel, O; Mesiti, G; Monteagudo, E; Pescatore, G; Rowley, M; Verdirame, M; Steinkühler, C A novel series of potent and selective ketone histone deacetylase inhibitors with antitumor activity in vivo. J Med Chem51:2350-3 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Histone deacetylase 4 [653-1084,H976Y] |
|---|
| Name: | Histone deacetylase 4 [653-1084,H976Y] |
|---|
| Synonyms: | HDAC4 | HDAC4_HUMAN | Histone Deacetylase 4 (HDAC4) Mutant (H976Y) | Histone deacetylase 4 gain of function (GOF) mutant | KIAA0288 |
|---|
| Type: | Catalytic domain |
|---|
| Mol. Mass.: | 46476.72 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | His-tagged HDAC4 GOF(gain of function, H976Y) catalytic domains (T653-L1084) was expressed in E.coli and purified by Nickel-Chelation affinity chromatography and anion exchange (MonoQ) chromatography. |
|---|
| Residue: | 432 |
|---|
| Sequence: | TTGLVYDTLMLKHQCTCGSSSSHPEHAGRIQSIWSRLQETGLRGKCECIRGRKATLEELQ
TVHSEAHTLLYGTNPLNRQKLDSKKLLGSLASVFVRLPCGGVGVDSDTIWNEVHSAGAAR
LAVGCVVELVFKVATGELKNGFAVVRPPGHHAEESTPMGFCYFNSVAVAAKLLQQRLSVS
KILIVDWDVHHGNGTQQAFYSDPSVLYMSLHRYDDGNFFPGSGAPDEVGTGPGVGFNVNM
AFTGGLDPPMGDAEYLAAFRTVVMPIASEFAPDVVLVSSGFDAVEGHPTPLGGYNLSARC
FGYLTKQLMGLAGGRIVLALEGGYDLTAICDASEACVSALLGNELDPLPEKVLQQRPNAN
AVRSMEKVMEIHSKYWRCLQRTTSTAGRSLIEAQTCENEEAETVTAMASLSVGVKPAEKR
PDEEPMEEEPPL
|
|
|
|---|
| BDBM19149 |
|---|
| BDBM25143 |
|---|
| Name | BDBM19149 |
|---|
| Synonyms: | CHEMBL98 | N-hydroxy-N'-phenyloctanediamide | SAHA | US10011611, SAHA | US10188756, Compound SAHA | US11207431, SAHA | US11505523, Compound SAHA | US9115116, SAHA | US9353061, SAHA | US9428447, SAHA | US9695181, Vorinostat | Vorinostat | Zolinza | cid_5311 | suberoylanilide hydroxamic acid |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H20N2O3 |
|---|
| Mol. Mass. | 264.3202 |
|---|
| SMILES | ONC(=O)CCCCCCC(=O)Nc1ccccc1 |
|---|
| Structure | 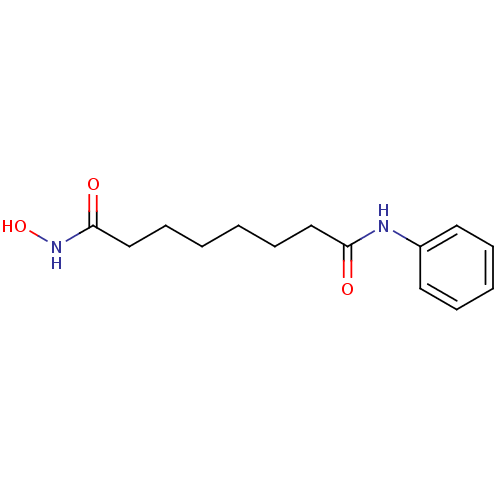
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Jones, P; Altamura, S; De Francesco, R; Paz, OG; Kinzel, O; Mesiti, G; Monteagudo, E; Pescatore, G; Rowley, M; Verdirame, M; Steinkühler, C A novel series of potent and selective ketone histone deacetylase inhibitors with antitumor activity in vivo. J Med Chem51:2350-3 (2008) [PubMed] Article
Jones, P; Altamura, S; De Francesco, R; Paz, OG; Kinzel, O; Mesiti, G; Monteagudo, E; Pescatore, G; Rowley, M; Verdirame, M; Steinkühler, C A novel series of potent and selective ketone histone deacetylase inhibitors with antitumor activity in vivo. J Med Chem51:2350-3 (2008) [PubMed] Article