| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Adenosylhomocysteine nucleosidase |
|---|
| Ligand | BDBM36435 |
|---|
| Substrate/Competitor | BDBM22111 |
|---|
| Meas. Tech. | Enzyme Activity Assay |
|---|
| pH | 7.5±0 |
|---|
| Temperature | 298.15±0 K |
|---|
| Kd | 10±0.0 nM |
|---|
| Citation |  Gutierrez, JA; Luo, M; Singh, V; Li, L; Brown, RL; Norris, GE; Evans, GB; Furneaux, RH; Tyler, PC; Painter, GF; Lenz, DH; Schramm, VL Picomolar inhibitors as transition-state probes of 5'-methylthioadenosine nucleosidases. ACS Chem Biol2:725-34 (2007) [PubMed] Article Gutierrez, JA; Luo, M; Singh, V; Li, L; Brown, RL; Norris, GE; Evans, GB; Furneaux, RH; Tyler, PC; Painter, GF; Lenz, DH; Schramm, VL Picomolar inhibitors as transition-state probes of 5'-methylthioadenosine nucleosidases. ACS Chem Biol2:725-34 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Adenosylhomocysteine nucleosidase |
|---|
| Name: | Adenosylhomocysteine nucleosidase |
|---|
| Synonyms: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | Methylthioadenosine Nucleosidase (MTAN) | mtnN |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 24666.14 |
|---|
| Organism: | Streptococcus pneumoniae |
|---|
| Description: | J0VBT9 |
|---|
| Residue: | 230 |
|---|
| Sequence: | MKIGIIAAMPEELAYLVQHLDNTQEQVVLGNTYHTGTIASHEVVLVESGIGKVMSAMSVA
ILADHFQVDALINTGSAGAVAEGIAVGDVVIADKLAYHDVDVTAFGYAYGQMAQQPLYFE
SDKTFVAQIQESLSQLDQNWHLGLIATGDSFVAGNDKIEAIKSHFPEVLAVEMEGAAIAQ
AAHALNLPVLVIRAMSDNANHEANIFFDEFIIEAGRRSAQVLLTFLKALD
|
|
|
|---|
| BDBM36435 |
|---|
| BDBM22111 |
|---|
| Name | BDBM36435 |
|---|
| Synonyms: | (3R,4S)-1-[(9-Deaza-adenin-9-yl)methyl]-4-ethylthio-methyl-3-hydroxypyrrolidine, 8 | CID11404039 | DADMe-ImmA-Et |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H21N5OS |
|---|
| Mol. Mass. | 307.414 |
|---|
| SMILES | CCSC[C@H]1CN(Cc2c[nH]c3c(N)ncnc23)C[C@@H]1O |
|---|
| Structure | 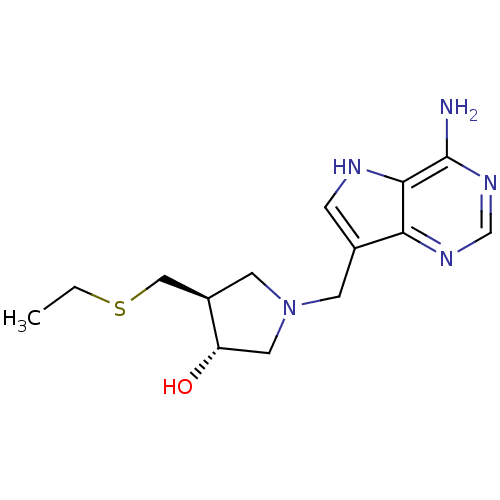
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Gutierrez, JA; Luo, M; Singh, V; Li, L; Brown, RL; Norris, GE; Evans, GB; Furneaux, RH; Tyler, PC; Painter, GF; Lenz, DH; Schramm, VL Picomolar inhibitors as transition-state probes of 5'-methylthioadenosine nucleosidases. ACS Chem Biol2:725-34 (2007) [PubMed] Article
Gutierrez, JA; Luo, M; Singh, V; Li, L; Brown, RL; Norris, GE; Evans, GB; Furneaux, RH; Tyler, PC; Painter, GF; Lenz, DH; Schramm, VL Picomolar inhibitors as transition-state probes of 5'-methylthioadenosine nucleosidases. ACS Chem Biol2:725-34 (2007) [PubMed] Article