| Reaction Details |
|---|
 | Report a problem with these data |
| Target | HIV-1 protease |
|---|
| Ligand | BDBM517 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Enzyme Inhibition Assay |
|---|
| Ki | 0.37±0.0 nM |
|---|
| Citation |  Chen, CA; Sieburth, SM; Glekas, A; Hewitt, GW; Trainor, GL; Erickson-Viitanen, S; Garber, SS; Cordova, B; Jeffry, S; Klabe, RM Drug design with a new transition state analog of the hydrated carbonyl: silicon-based inhibitors of the HIV protease. Chem Biol8:1161-6 (2001) [PubMed] Article Chen, CA; Sieburth, SM; Glekas, A; Hewitt, GW; Trainor, GL; Erickson-Viitanen, S; Garber, SS; Cordova, B; Jeffry, S; Klabe, RM Drug design with a new transition state analog of the hydrated carbonyl: silicon-based inhibitors of the HIV protease. Chem Biol8:1161-6 (2001) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| HIV-1 protease |
|---|
| Name: | HIV-1 protease |
|---|
| Synonyms: | HIV-1 protease wild type |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 10757.68 |
|---|
| Organism: | Human immunodeficiency virus |
|---|
| Description: | O90785 |
|---|
| Residue: | 99 |
|---|
| Sequence: | PQITLWQRPLVTVKIGGQLREALLDTGADDTVLEDINLPGKWKPKMIGGIGGFIKVKQYE
QVLIEICGKKAIGTVLVGPTPVNIIGRNMLTQIGCTLNF
|
|
|
|---|
| BDBM517 |
|---|
| n/a |
|---|
| Name | BDBM517 |
|---|
| Synonyms: | (2S)-1-[(2S,4R)-4-benzyl-2-hydroxy-4-{[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]carbamoyl}butyl]-N-tert-butyl-4-(pyridin-3-ylmethyl)piperazine-2-carboxamide | CHEMBL115 | Crixivan | INDINAVIR SULFATE | Indinavir | Indinavir, 19 | L-735, 524 | MK639 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C36H47N5O4 |
|---|
| Mol. Mass. | 613.7895 |
|---|
| SMILES | CC(C)(C)NC(=O)[C@@H]1CN(Cc2cccnc2)CCN1C[C@@H](O)C[C@@H](Cc1ccccc1)C(=O)N[C@@H]1[C@H](O)Cc2ccccc12 |@:19,@@:9| |
|---|
| Structure | 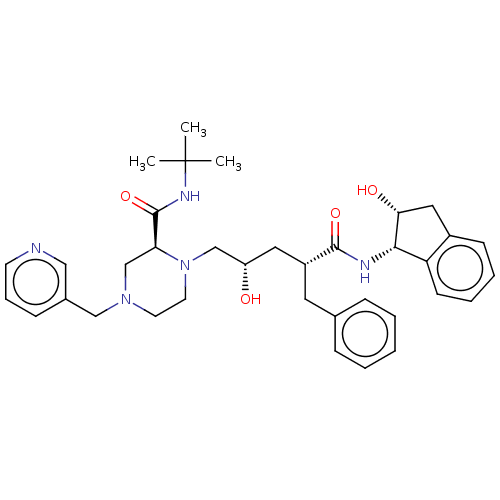
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Chen, CA; Sieburth, SM; Glekas, A; Hewitt, GW; Trainor, GL; Erickson-Viitanen, S; Garber, SS; Cordova, B; Jeffry, S; Klabe, RM Drug design with a new transition state analog of the hydrated carbonyl: silicon-based inhibitors of the HIV protease. Chem Biol8:1161-6 (2001) [PubMed] Article
Chen, CA; Sieburth, SM; Glekas, A; Hewitt, GW; Trainor, GL; Erickson-Viitanen, S; Garber, SS; Cordova, B; Jeffry, S; Klabe, RM Drug design with a new transition state analog of the hydrated carbonyl: silicon-based inhibitors of the HIV protease. Chem Biol8:1161-6 (2001) [PubMed] Article