| Reaction Details |
|---|
 | Report a problem with these data |
| Target | S-ribosylhomocysteine lyase |
|---|
| Ligand | BDBM65976 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Absorbance Microorganism-Based Dose Retest to Identify Inhibitors of Vibrio harveyi |
|---|
| EC50 | >160000±n/a nM |
|---|
| Citation |  PubChem, PC Absorbance Microorganism-Based Dose Retest to Identify Inhibitors of Vibrio harveyi PubChem Bioassay(2010)[AID] PubChem, PC Absorbance Microorganism-Based Dose Retest to Identify Inhibitors of Vibrio harveyi PubChem Bioassay(2010)[AID] |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| S-ribosylhomocysteine lyase |
|---|
| Name: | S-ribosylhomocysteine lyase |
|---|
| Synonyms: | LuxS |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 15564.06 |
|---|
| Organism: | Vibrio harveyi |
|---|
| Description: | B0LUE4 |
|---|
| Residue: | 140 |
|---|
| Sequence: | MPLLDSFTVDHTRMNAPAVRVAKTMQTPKGDTITVFDLRFTAPNKDILSEKGIHTLEHLY
AGFMRNHLNGDSVEIIDISPMGCRTGFYMSLIGTPSEQQVADAWIAAMEDVLKVENQNKI
PELNEYQCGTAAMHSLDEAK
|
|
|
|---|
| BDBM65976 |
|---|
| n/a |
|---|
| Name | BDBM65976 |
|---|
| Synonyms: | MLS000099320 | N-(6-methyl-2-pyridinyl)-4-phenoxybenzenesulfonamide | N-(6-methyl-2-pyridyl)-4-phenoxy-benzenesulfonamide | N-(6-methylpyridin-2-yl)-4-phenoxy-benzenesulfonamide | N-(6-methylpyridin-2-yl)-4-phenoxybenzenesulfonamide | SMR000071466 | cid_801216 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H16N2O3S |
|---|
| Mol. Mass. | 340.396 |
|---|
| SMILES | Cc1cccc(NS(=O)(=O)c2ccc(Oc3ccccc3)cc2)n1 |
|---|
| Structure | 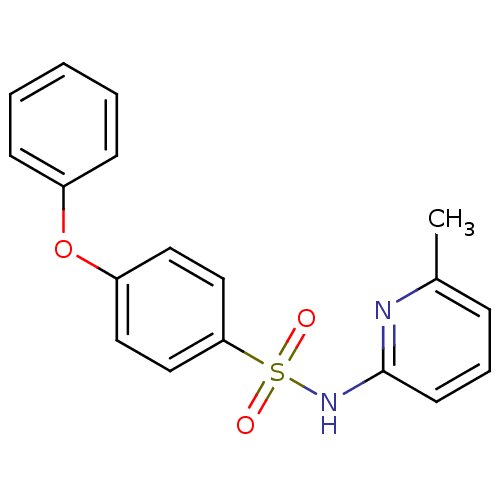
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Absorbance Microorganism-Based Dose Retest to Identify Inhibitors of Vibrio harveyi PubChem Bioassay(2010)[AID]
PubChem, PC Absorbance Microorganism-Based Dose Retest to Identify Inhibitors of Vibrio harveyi PubChem Bioassay(2010)[AID]