| Reaction Details |
|---|
 | Report a problem with these data |
| Target | cAMP-specific 3',5'-cyclic phosphodiesterase 7B |
|---|
| Ligand | BDBM50258898 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_1690781 |
|---|
| IC50 | >10000±n/a nM |
|---|
| Citation |  Mikami, S; Nakamura, S; Ashizawa, T; Nomura, I; Kawasaki, M; Sasaki, S; Oki, H; Kokubo, H; Hoffman, ID; Zou, H; Uchiyama, N; Nakashima, K; Kamiguchi, N; Imada, H; Suzuki, N; Iwashita, H; Taniguchi, T Discovery of Clinical Candidate N-((1S)-1-(3-Fluoro-4-(trifluoromethoxy)phenyl)-2-methoxyethyl)-7-methoxy-2-oxo-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide (TAK-915): A Highly Potent, Selective, and Brain-Penetrating Phosphodiesterase 2A Inhibitor for the Treatment of Cognitive Disorders. J Med Chem60:7677-7702 (2017) [PubMed] Article Mikami, S; Nakamura, S; Ashizawa, T; Nomura, I; Kawasaki, M; Sasaki, S; Oki, H; Kokubo, H; Hoffman, ID; Zou, H; Uchiyama, N; Nakashima, K; Kamiguchi, N; Imada, H; Suzuki, N; Iwashita, H; Taniguchi, T Discovery of Clinical Candidate N-((1S)-1-(3-Fluoro-4-(trifluoromethoxy)phenyl)-2-methoxyethyl)-7-methoxy-2-oxo-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide (TAK-915): A Highly Potent, Selective, and Brain-Penetrating Phosphodiesterase 2A Inhibitor for the Treatment of Cognitive Disorders. J Med Chem60:7677-7702 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| cAMP-specific 3',5'-cyclic phosphodiesterase 7B |
|---|
| Name: | cAMP-specific 3',5'-cyclic phosphodiesterase 7B |
|---|
| Synonyms: | 3',5'-cyclic phosphodiesterase | PDE7B | PDE7B_HUMAN | Phosphodiesterase 7B | Phosphodiesterase 7B (PDE7B) | cAMP-specific 3',5'-cyclic phosphodiesterase 7B |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 51842.76 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9NP56 |
|---|
| Residue: | 450 |
|---|
| Sequence: | MSCLMVERCGEILFENPDQNAKCVCMLGDIRLRGQTGVRAERRGSYPFIDFRLLNSTTYS
GEIGTKKKVKRLLSFQRYFHASRLLRGIIPQAPLHLLDEDYLGQARHMLSKVGMWDFDIF
LFDRLTNGNSLVTLLCHLFNTHGLIHHFKLDMVTLHRFLVMVQEDYHSQNPYHNAVHAAD
VTQAMHCYLKEPKLASFLTPLDIMLGLLAAAAHDVDHPGVNQPFLIKTNHHLANLYQNMS
VLENHHWRSTIGMLRESRLLAHLPKEMTQDIEQQLGSLILATDINRQNEFLTRLKAHLHN
KDLRLEDAQDRHFMLQIALKCADICNPCRIWEMSKQWSERVCEEFYRQGELEQKFELEIS
PLCNQQKDSIPSIQIGFMSYIVEPLFREWAHFTGNSTLSENMLGHLAHNKAQWKSLLPRQ
HRSRGSSGSGPDHDHAGQGTESEEQEGDSP
|
|
|
|---|
| BDBM50258898 |
|---|
| n/a |
|---|
| Name | BDBM50258898 |
|---|
| Synonyms: | CHEMBL4064344 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H17F3N4O3 |
|---|
| Mol. Mass. | 394.3478 |
|---|
| SMILES | CCC(NC(=O)N1CC(=O)Nc2cccnc12)c1ccc(OC(F)(F)F)cc1 |
|---|
| Structure | 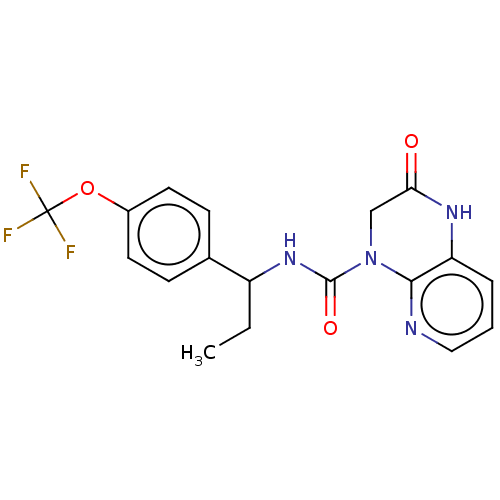
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Mikami, S; Nakamura, S; Ashizawa, T; Nomura, I; Kawasaki, M; Sasaki, S; Oki, H; Kokubo, H; Hoffman, ID; Zou, H; Uchiyama, N; Nakashima, K; Kamiguchi, N; Imada, H; Suzuki, N; Iwashita, H; Taniguchi, T Discovery of Clinical Candidate N-((1S)-1-(3-Fluoro-4-(trifluoromethoxy)phenyl)-2-methoxyethyl)-7-methoxy-2-oxo-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide (TAK-915): A Highly Potent, Selective, and Brain-Penetrating Phosphodiesterase 2A Inhibitor for the Treatment of Cognitive Disorders. J Med Chem60:7677-7702 (2017) [PubMed] Article
Mikami, S; Nakamura, S; Ashizawa, T; Nomura, I; Kawasaki, M; Sasaki, S; Oki, H; Kokubo, H; Hoffman, ID; Zou, H; Uchiyama, N; Nakashima, K; Kamiguchi, N; Imada, H; Suzuki, N; Iwashita, H; Taniguchi, T Discovery of Clinical Candidate N-((1S)-1-(3-Fluoro-4-(trifluoromethoxy)phenyl)-2-methoxyethyl)-7-methoxy-2-oxo-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide (TAK-915): A Highly Potent, Selective, and Brain-Penetrating Phosphodiesterase 2A Inhibitor for the Treatment of Cognitive Disorders. J Med Chem60:7677-7702 (2017) [PubMed] Article