

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | cGMP-dependent 3',5'-cyclic phosphodiesterase | ||
| Ligand | BDBM50263752 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1698247 (CHEMBL4049137) | ||
| IC50 | 0.230000±n/a nM | ||
| Citation |  Obach, RS; Walker, GS; Sharma, R; Jenkinson, S; Tran, TP; Stepan, AF Lead Diversification at the Nanomole Scale Using Liver Microsomes and Quantitative Nuclear Magnetic Resonance Spectroscopy: Application to Phosphodiesterase 2 Inhibitors. J Med Chem61:3626-3640 (2018) [PubMed] Article Obach, RS; Walker, GS; Sharma, R; Jenkinson, S; Tran, TP; Stepan, AF Lead Diversification at the Nanomole Scale Using Liver Microsomes and Quantitative Nuclear Magnetic Resonance Spectroscopy: Application to Phosphodiesterase 2 Inhibitors. J Med Chem61:3626-3640 (2018) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| cGMP-dependent 3',5'-cyclic phosphodiesterase | |||
| Name: | cGMP-dependent 3',5'-cyclic phosphodiesterase | ||
| Synonyms: | CGS-PDE | Cyclic GMP-stimulated phosphodiesterase | Homo sapiens phosphodiesterase 2A (PDE2A) | NM_002599 | PDE2A | PDE2A_HUMAN | cGSPDE | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 105691.58 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | O00408 | ||
| Residue: | 941 | ||
| Sequence: |
| ||
| BDBM50263752 | |||
| n/a | |||
| Name | BDBM50263752 | ||
| Synonyms: | CHEMBL4092829 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C18H13F5N6O | ||
| Mol. Mass. | 424.3274 | ||
| SMILES | Cc1n[nH]c(c1-c1nc(C(F)F)c2n1nc(C)[nH]c2=O)-c1ccc(cc1)C(F)(F)F |(26.88,-15.97,;25.65,-16.89,;25.67,-18.43,;24.22,-18.93,;23.29,-17.7,;24.18,-16.44,;23.68,-14.98,;24.56,-13.72,;23.64,-12.49,;24.09,-11.02,;23.1,-9.84,;25.6,-10.75,;22.18,-12.99,;22.2,-14.53,;20.88,-15.33,;19.54,-14.58,;18.22,-15.37,;19.51,-13.03,;20.83,-12.25,;20.81,-10.71,;21.75,-17.73,;21.01,-19.07,;19.47,-19.1,;18.67,-17.78,;19.42,-16.43,;20.96,-16.41,;17.13,-17.8,;16.72,-19.28,;15.64,-18.19,;16.34,-16.49,)| | ||
| Structure | 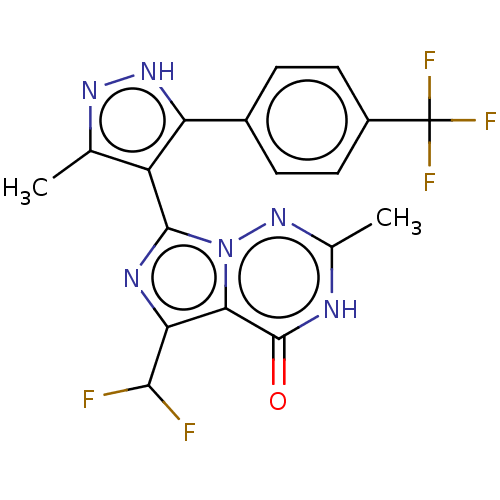
| ||