

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Neuronal acetylcholine receptor subunit alpha-7 | ||
| Ligand | BDBM50451606 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1745955 (CHEMBL4180465) | ||
| EC50 | 13±n/a nM | ||
| Citation |  Cook, J; Zusi, FC; Hill, MD; Fang, H; Pearce, B; Park, H; Gallagher, L; McDonald, IM; Bristow, L; Macor, JE; Olson, RE Design and synthesis of a novel series of (1'S,2R,4'S)-3H-4'-azaspiro[benzo[4,5]imidazo[2,1-b]oxazole-2,2'-bicyclo[2.2.2]octanes] with high affinity for the ?7 neuronal nicotinic receptor. Bioorg Med Chem Lett27:5002-5005 (2017) [PubMed] Article Cook, J; Zusi, FC; Hill, MD; Fang, H; Pearce, B; Park, H; Gallagher, L; McDonald, IM; Bristow, L; Macor, JE; Olson, RE Design and synthesis of a novel series of (1'S,2R,4'S)-3H-4'-azaspiro[benzo[4,5]imidazo[2,1-b]oxazole-2,2'-bicyclo[2.2.2]octanes] with high affinity for the ?7 neuronal nicotinic receptor. Bioorg Med Chem Lett27:5002-5005 (2017) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Neuronal acetylcholine receptor subunit alpha-7 | |||
| Name: | Neuronal acetylcholine receptor subunit alpha-7 | ||
| Synonyms: | ACHA7_RAT | Acra7 | Cholinergic, Nicotinic Alpha7 | Cholinergic, Nicotinic Alpha7/5-HT3 | Chrna7 | Neuronal acetylcholine receptor | Neuronal acetylcholine receptor (alpha7 nAChR) | Neuronal acetylcholine receptor subunit alpha 7 | Neuronal acetylcholine receptor subunit alpha-7 | Neuronal acetylcholine receptor subunit alpha-7 (nAChR alpha7) | Neuronal acetylcholine receptor subunit alpha-7 (nAChR) | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 56502.44 | ||
| Organism: | Rattus norvegicus (Rat) | ||
| Description: | Q05941 | ||
| Residue: | 502 | ||
| Sequence: |
| ||
| BDBM50451606 | |||
| n/a | |||
| Name | BDBM50451606 | ||
| Synonyms: | CHEMBL4214778 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C16H16F3N3O | ||
| Mol. Mass. | 323.3129 | ||
| SMILES | FC(F)(F)c1ccc2n3C[C@@]4(CN5CCC4CC5)Oc3nc2c1 |r,wU:10.19,THB:18:10:13.14:17.16,(24.67,-19.03,;25.09,-20.6,;26.24,-19.45,;26.25,-21.75,;23.54,-21.04,;23.14,-22.53,;21.66,-22.93,;20.57,-21.85,;19.03,-21.94,;17.84,-22.9,;16.55,-22.07,;16.85,-23.54,;15.41,-22.87,;13.8,-23.57,;13.59,-22.11,;15.14,-21.44,;15.2,-19.72,;15.68,-20.88,;16.94,-20.59,;18.47,-20.5,;19.67,-19.53,;20.95,-20.36,;22.44,-19.95,)| | ||
| Structure | 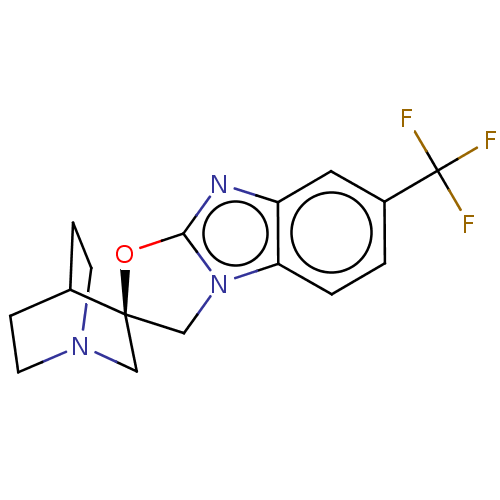
| ||