| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Nonstructural protein 5A |
|---|
| Ligand | BDBM50453100 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1749042 (CHEMBL4183552) |
|---|
| EC50 | 0.064±n/a nM |
|---|
| Citation |  Wagner, R; Randolph, JT; Patel, SV; Nelson, L; Matulenko, MA; Keddy, R; Pratt, JK; Liu, D; Krueger, AC; Donner, PL; Hutchinson, DK; Flentge, C; Betebenner, D; Rockway, T; Maring, CJ; Ng, TI; Krishnan, P; Pilot-Matias, T; Collins, C; Panchal, N; Reisch, T; Dekhtyar, T; Mondal, R; Stolarik, DF; Gao, Y; Gao, W; Beno, DA; Kati, WM Highlights of the Structure-Activity Relationships of Benzimidazole Linked Pyrrolidines Leading to the Discovery of the Hepatitis C Virus NS5A Inhibitor Pibrentasvir (ABT-530). J Med Chem61:4052-4066 (2018) [PubMed] Article Wagner, R; Randolph, JT; Patel, SV; Nelson, L; Matulenko, MA; Keddy, R; Pratt, JK; Liu, D; Krueger, AC; Donner, PL; Hutchinson, DK; Flentge, C; Betebenner, D; Rockway, T; Maring, CJ; Ng, TI; Krishnan, P; Pilot-Matias, T; Collins, C; Panchal, N; Reisch, T; Dekhtyar, T; Mondal, R; Stolarik, DF; Gao, Y; Gao, W; Beno, DA; Kati, WM Highlights of the Structure-Activity Relationships of Benzimidazole Linked Pyrrolidines Leading to the Discovery of the Hepatitis C Virus NS5A Inhibitor Pibrentasvir (ABT-530). J Med Chem61:4052-4066 (2018) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Nonstructural protein 5A |
|---|
| Name: | Nonstructural protein 5A |
|---|
| Synonyms: | NS5A | Nonstructural protein 5A |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 48587.20 |
|---|
| Organism: | Hepatitis C virus |
|---|
| Description: | ChEMBL_108438 |
|---|
| Residue: | 447 |
|---|
| Sequence: | SGSWLRDVWDWICTVLTDFKTWLQSKLLPRIPGVPFLSCQRGYKGVWRGDGIMHTTCPCG
AQITGHVKNGSMRIVGPKTCSNTWHGTFPINTYTTGPCTPSPAPNYSRALWRVAAEEYVE
VTRVGDFHYVTGMTTDNVKCPCQVPAPEFFTEVDGVRLHRYAPACKPLLREEVTFMVGLN
QYLVGSQLPCEPEPDVTVLTSMLTDPSHITAETAGRRLARGSPPSLASSSASQLSAPSLK
ATCTTRHDSPDADLIEANLLWRQEMGGNITRVESENKVVILDSFDPLQAEEDEREVSVPA
EILRKSRKFPRAMPIWARPDYNPPLLESWKDPDYVPPVVHGCPLPPAKAPPIPPPRRKRT
VVLTESTVSSALAELATKTFGSSGSSAVDSGTATAPPDQLPGDGDSGSDVESYSSMPPLE
GEPGDPDLSDGSWSTMSEEASEDVVCC
|
|
|
|---|
| BDBM50453100 |
|---|
| n/a |
|---|
| Name | BDBM50453100 |
|---|
| Synonyms: | A-1325912.0 | ABT-530 | Pibrentasvir |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C57H65F5N10O8 |
|---|
| Mol. Mass. | 1113.1802 |
|---|
| SMILES | CO[C@H](C)[C@H](NC(=O)OC)C(=O)N1CCC[C@H]1c1nc2cc([C@H]3CC[C@@H](N3c3cc(F)c(N4CCC(CC4)c4ccc(F)cc4)c(F)c3)c3cc4nc([nH]c4cc3F)[C@@H]3CCCN3C(=O)[C@@H](NC(=O)OC)[C@@H](C)OC)c(F)cc2[nH]1 |r| |
|---|
| Structure | 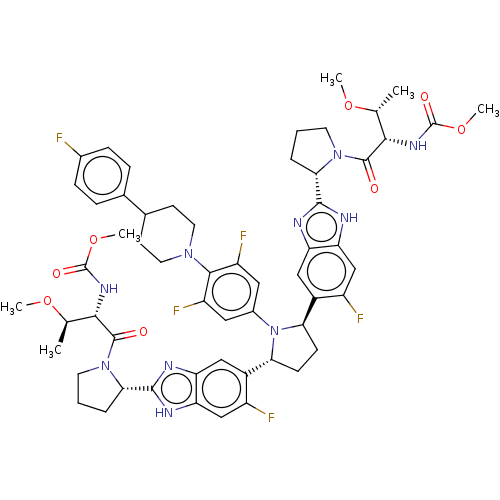
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wagner, R; Randolph, JT; Patel, SV; Nelson, L; Matulenko, MA; Keddy, R; Pratt, JK; Liu, D; Krueger, AC; Donner, PL; Hutchinson, DK; Flentge, C; Betebenner, D; Rockway, T; Maring, CJ; Ng, TI; Krishnan, P; Pilot-Matias, T; Collins, C; Panchal, N; Reisch, T; Dekhtyar, T; Mondal, R; Stolarik, DF; Gao, Y; Gao, W; Beno, DA; Kati, WM Highlights of the Structure-Activity Relationships of Benzimidazole Linked Pyrrolidines Leading to the Discovery of the Hepatitis C Virus NS5A Inhibitor Pibrentasvir (ABT-530). J Med Chem61:4052-4066 (2018) [PubMed] Article
Wagner, R; Randolph, JT; Patel, SV; Nelson, L; Matulenko, MA; Keddy, R; Pratt, JK; Liu, D; Krueger, AC; Donner, PL; Hutchinson, DK; Flentge, C; Betebenner, D; Rockway, T; Maring, CJ; Ng, TI; Krishnan, P; Pilot-Matias, T; Collins, C; Panchal, N; Reisch, T; Dekhtyar, T; Mondal, R; Stolarik, DF; Gao, Y; Gao, W; Beno, DA; Kati, WM Highlights of the Structure-Activity Relationships of Benzimidazole Linked Pyrrolidines Leading to the Discovery of the Hepatitis C Virus NS5A Inhibitor Pibrentasvir (ABT-530). J Med Chem61:4052-4066 (2018) [PubMed] Article