| Reaction Details |
|---|
 | Report a problem with these data |
| Target | UDP-glucuronosyltransferase 1A1 |
|---|
| Ligand | BDBM50460863 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1770476 (CHEMBL4222588) |
|---|
| IC50 | 1000±n/a nM |
|---|
| Citation |  Ginnetti, AT; Paone, DV; Stauffer, SR; Potteiger, CM; Shaw, AW; Deng, J; Mulhearn, JJ; Nguyen, DN; Segerdell, C; Anquandah, J; Calamari, A; Cheng, G; Leitl, MD; Liang, A; Moore, E; Panigel, J; Urban, M; Wang, J; Fillgrove, K; Tang, C; Cook, S; Kane, S; Salvatore, CA; Graham, SL; Burgey, CS Identification of second-generation P2X3 antagonists for treatment of pain. Bioorg Med Chem Lett28:1392-1396 (2018) [PubMed] Article Ginnetti, AT; Paone, DV; Stauffer, SR; Potteiger, CM; Shaw, AW; Deng, J; Mulhearn, JJ; Nguyen, DN; Segerdell, C; Anquandah, J; Calamari, A; Cheng, G; Leitl, MD; Liang, A; Moore, E; Panigel, J; Urban, M; Wang, J; Fillgrove, K; Tang, C; Cook, S; Kane, S; Salvatore, CA; Graham, SL; Burgey, CS Identification of second-generation P2X3 antagonists for treatment of pain. Bioorg Med Chem Lett28:1392-1396 (2018) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| UDP-glucuronosyltransferase 1A1 |
|---|
| Name: | UDP-glucuronosyltransferase 1A1 |
|---|
| Synonyms: | Bilirubin-specific UDPGT isozyme 1 | GNT1 | UD11_HUMAN | UDP-glucuronosyltransferase 1-1 | UDP-glucuronosyltransferase 1-A | UDP-glucuronosyltransferase 1A1 | UDPGT 1-1 | UGT-1A | UGT1 | UGT1*1 | UGT1-01 | UGT1.1 | UGT1A | UGT1A1 | Uridine-5'-diphosphoglucuronosyltransferase 1A1 | hUG-BR1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 59604.34 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P22309 |
|---|
| Residue: | 533 |
|---|
| Sequence: | MAVESQGGRPLVLGLLLCVLGPVVSHAGKILLIPVDGSHWLSMLGAIQQLQQRGHEIVVL
APDASLYIRDGAFYTLKTYPVPFQREDVKESFVSLGHNVFENDSFLQRVIKTYKKIKKDS
AMLLSGCSHLLHNKELMASLAESSFDVMLTDPFLPCSPIVAQYLSLPTVFFLHALPCSLE
FEATQCPNPFSYVPRPLSSHSDHMTFLQRVKNMLIAFSQNFLCDVVYSPYATLASEFLQR
EVTVQDLLSSASVWLFRSDFVKDYPRPIMPNMVFVGGINCLHQNPLSQEFEAYINASGEH
GIVVFSLGSMVSEIPEKKAMAIADALGKIPQTVLWRYTGTRPSNLANNTILVKWLPQNDL
LGHPMTRAFITHAGSHGVYESICNGVPMVMMPLFGDQMDNAKRMETKGAGVTLNVLEMTS
EDLENALKAVINDKSYKENIMRLSSLHKDRPVEPLDLAVFWVEFVMRHKGAPHLRPAAHD
LTWYQYHSLDVIGFLLAVVLTVAFITFKCCAYGYRKCLGKKGRVKKAHKSKTH
|
|
|
|---|
| BDBM50460863 |
|---|
| n/a |
|---|
| Name | BDBM50460863 |
|---|
| Synonyms: | CHEMBL4227228 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C28H24FN5O2 |
|---|
| Mol. Mass. | 481.5209 |
|---|
| SMILES | C[C@@H](NC(=O)c1cc(cc(c1)-c1ccc(C)cn1)C1=NO[C@@H](C1)c1ccccn1)c1ccc(F)cn1 |r,t:20| |
|---|
| Structure | 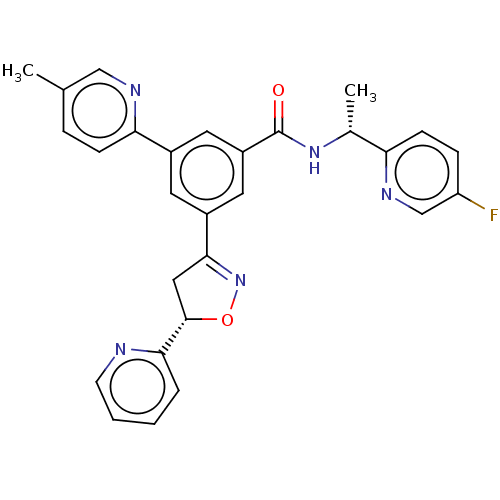
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ginnetti, AT; Paone, DV; Stauffer, SR; Potteiger, CM; Shaw, AW; Deng, J; Mulhearn, JJ; Nguyen, DN; Segerdell, C; Anquandah, J; Calamari, A; Cheng, G; Leitl, MD; Liang, A; Moore, E; Panigel, J; Urban, M; Wang, J; Fillgrove, K; Tang, C; Cook, S; Kane, S; Salvatore, CA; Graham, SL; Burgey, CS Identification of second-generation P2X3 antagonists for treatment of pain. Bioorg Med Chem Lett28:1392-1396 (2018) [PubMed] Article
Ginnetti, AT; Paone, DV; Stauffer, SR; Potteiger, CM; Shaw, AW; Deng, J; Mulhearn, JJ; Nguyen, DN; Segerdell, C; Anquandah, J; Calamari, A; Cheng, G; Leitl, MD; Liang, A; Moore, E; Panigel, J; Urban, M; Wang, J; Fillgrove, K; Tang, C; Cook, S; Kane, S; Salvatore, CA; Graham, SL; Burgey, CS Identification of second-generation P2X3 antagonists for treatment of pain. Bioorg Med Chem Lett28:1392-1396 (2018) [PubMed] Article