| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 3 |
|---|
| Ligand | BDBM50105327 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1779294 (CHEMBL4236286) |
|---|
| Ki | 4.3±n/a nM |
|---|
| Citation |  Fournier, JF; Bhurruth-Alcor, Y; Musicki, B; Aubert, J; Aurelly, M; Bouix-Peter, C; Bouquet, K; Chantalat, L; Delorme, M; Drean, B; Duvert, G; Fleury-Bregeot, N; Gauthier, B; Grisendi, K; Harris, CS; Hennequin, LF; Isabet, T; Joly, F; Lafitte, G; Millois, C; Morgentin, R; Pascau, J; Piwnica, D; Rival, Y; Soulet, C; Thoreau, É; Tomas, L Squaramides as novel class I and IIB histone deacetylase inhibitors for topical treatment of cutaneous t-cell lymphoma. Bioorg Med Chem Lett28:2985-2992 (2018) [PubMed] Article Fournier, JF; Bhurruth-Alcor, Y; Musicki, B; Aubert, J; Aurelly, M; Bouix-Peter, C; Bouquet, K; Chantalat, L; Delorme, M; Drean, B; Duvert, G; Fleury-Bregeot, N; Gauthier, B; Grisendi, K; Harris, CS; Hennequin, LF; Isabet, T; Joly, F; Lafitte, G; Millois, C; Morgentin, R; Pascau, J; Piwnica, D; Rival, Y; Soulet, C; Thoreau, É; Tomas, L Squaramides as novel class I and IIB histone deacetylase inhibitors for topical treatment of cutaneous t-cell lymphoma. Bioorg Med Chem Lett28:2985-2992 (2018) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone deacetylase 3 |
|---|
| Name: | Histone deacetylase 3 |
|---|
| Synonyms: | HD3 | HDAC3 | HDAC3_HUMAN | Histone deacetylase 3 (HDAC3) | Human HDAC3 | RPD3-2 | SMAP45 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 48829.55 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O15379 |
|---|
| Residue: | 428 |
|---|
| Sequence: | MAKTVAYFYDPDVGNFHYGAGHPMKPHRLALTHSLVLHYGLYKKMIVFKPYQASQHDMCR
FHSEDYIDFLQRVSPTNMQGFTKSLNAFNVGDDCPVFPGLFEFCSRYTGASLQGATQLNN
KICDIAINWAGGLHHAKKFEASGFCYVNDIVIGILELLKYHPRVLYIDIDIHHGDGVQEA
FYLTDRVMTVSFHKYGNYFFPGTGDMYEVGAESGRYYCLNVPLRDGIDDQSYKHLFQPVI
NQVVDFYQPTCIVLQCGADSLGCDRLGCFNLSIRGHGECVEYVKSFNIPLLVLGGGGYTV
RNVARCWTYETSLLVEEAISEELPYSEYFEYFAPDFTLHPDVSTRIENQNSRQYLDQIRQ
TIFENLKMLNHAPSVQIHDVPADLLTYDRTDEADAEERGPEENYSRPEAPNEFYDGDHDN
DKESDVEI
|
|
|
|---|
| BDBM50105327 |
|---|
| n/a |
|---|
| Name | BDBM50105327 |
|---|
| Synonyms: | JNJ-26481585 | Quisinostat |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H26N6O2 |
|---|
| Mol. Mass. | 394.4701 |
|---|
| SMILES | Cn1cc(CNCC2CCN(CC2)c2ncc(cn2)C(=O)NO)c2ccccc12 |
|---|
| Structure | 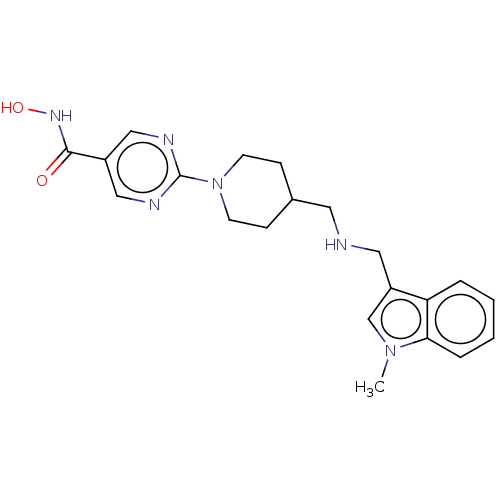
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Fournier, JF; Bhurruth-Alcor, Y; Musicki, B; Aubert, J; Aurelly, M; Bouix-Peter, C; Bouquet, K; Chantalat, L; Delorme, M; Drean, B; Duvert, G; Fleury-Bregeot, N; Gauthier, B; Grisendi, K; Harris, CS; Hennequin, LF; Isabet, T; Joly, F; Lafitte, G; Millois, C; Morgentin, R; Pascau, J; Piwnica, D; Rival, Y; Soulet, C; Thoreau, É; Tomas, L Squaramides as novel class I and IIB histone deacetylase inhibitors for topical treatment of cutaneous t-cell lymphoma. Bioorg Med Chem Lett28:2985-2992 (2018) [PubMed] Article
Fournier, JF; Bhurruth-Alcor, Y; Musicki, B; Aubert, J; Aurelly, M; Bouix-Peter, C; Bouquet, K; Chantalat, L; Delorme, M; Drean, B; Duvert, G; Fleury-Bregeot, N; Gauthier, B; Grisendi, K; Harris, CS; Hennequin, LF; Isabet, T; Joly, F; Lafitte, G; Millois, C; Morgentin, R; Pascau, J; Piwnica, D; Rival, Y; Soulet, C; Thoreau, É; Tomas, L Squaramides as novel class I and IIB histone deacetylase inhibitors for topical treatment of cutaneous t-cell lymphoma. Bioorg Med Chem Lett28:2985-2992 (2018) [PubMed] Article