| Reaction Details |
|---|
 | Report a problem with these data |
| Target | E3 ubiquitin-protein ligase XIAP |
|---|
| Ligand | BDBM18073 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1800813 (CHEMBL4273105) |
|---|
| IC50 | 80200±n/a nM |
|---|
| Citation |  Channar, PA; Afzal, S; Ejaz, SA; Saeed, A; Larik, FA; Mahesar, PA; Lecka, J; Sévigny, J; Erben, MF; Iqbal, J Exploration of carboxy pyrazole derivatives: Synthesis, alkaline phosphatase, nucleotide pyrophosphatase/phosphodiesterase and nucleoside triphosphate diphosphohydrolase inhibition studies with potential anticancer profile. Eur J Med Chem156:461-478 (2018) [PubMed] Article Channar, PA; Afzal, S; Ejaz, SA; Saeed, A; Larik, FA; Mahesar, PA; Lecka, J; Sévigny, J; Erben, MF; Iqbal, J Exploration of carboxy pyrazole derivatives: Synthesis, alkaline phosphatase, nucleotide pyrophosphatase/phosphodiesterase and nucleoside triphosphate diphosphohydrolase inhibition studies with potential anticancer profile. Eur J Med Chem156:461-478 (2018) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| E3 ubiquitin-protein ligase XIAP |
|---|
| Name: | E3 ubiquitin-protein ligase XIAP |
|---|
| Synonyms: | API3 | BIRC4 | E3 ubiquitin-protein ligase XIAP | IAP3 | Inhibitor of apoptosis protein 3 | Inhibitor of apoptosis protein 3 (XIAP) | X-linked inhibitor of apoptosis | X-linked inhibitor of apoptosis protein (XIAP) | XIAP | XIAP_HUMAN |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 56685.27 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P98170 |
|---|
| Residue: | 497 |
|---|
| Sequence: | MTFNSFEGSKTCVPADINKEEEFVEEFNRLKTFANFPSGSPVSASTLARAGFLYTGEGDT
VRCFSCHAAVDRWQYGDSAVGRHRKVSPNCRFINGFYLENSATQSTNSGIQNGQYKVENY
LGSRDHFALDRPSETHADYLLRTGQVVDISDTIYPRNPAMYSEEARLKSFQNWPDYAHLT
PRELASAGLYYTGIGDQVQCFCCGGKLKNWEPCDRAWSEHRRHFPNCFFVLGRNLNIRSE
SDAVSSDRNFPNSTNLPRNPSMADYEARIFTFGTWIYSVNKEQLARAGFYALGEGDKVKC
FHCGGGLTDWKPSEDPWEQHAKWYPGCKYLLEQKGQEYINNIHLTHSLEECLVRTTEKTP
SLTRRIDDTIFQNPMVQEAIRMGFSFKDIKKIMEEKIQISGSNYKSLEVLVADLVNAQKD
SMQDESSQTSLQKEISTEEQLRRLQEEKLCKICMDRNIAIVFVPCGHLVTCKQCAEAVDK
CPMCYTVITFKQKIFMS
|
|
|
|---|
| BDBM18073 |
|---|
| n/a |
|---|
| Name | BDBM18073 |
|---|
| Synonyms: | (2S)-2-amino-3-phenylpropanoic acid | CHEMBL301523 | L-[2,3,4,5,6-3H]phenylalanine | L-phenylalanine | Phenylalanine | US11021454, Compound L-phe |
|---|
| Type | Amino Acid |
|---|
| Emp. Form. | C9H11NO2 |
|---|
| Mol. Mass. | 165.1891 |
|---|
| SMILES | N[C@@H](Cc1ccccc1)C(O)=O |
|---|
| Structure | 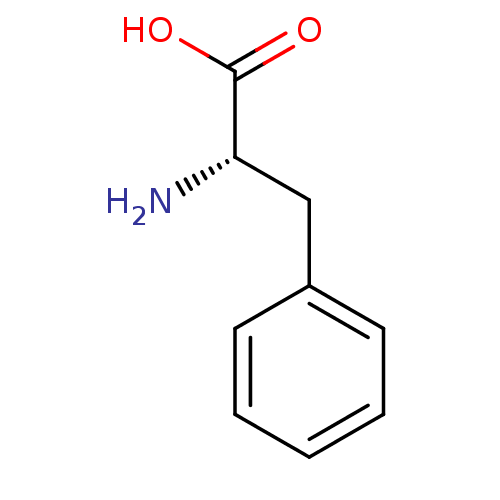
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Channar, PA; Afzal, S; Ejaz, SA; Saeed, A; Larik, FA; Mahesar, PA; Lecka, J; Sévigny, J; Erben, MF; Iqbal, J Exploration of carboxy pyrazole derivatives: Synthesis, alkaline phosphatase, nucleotide pyrophosphatase/phosphodiesterase and nucleoside triphosphate diphosphohydrolase inhibition studies with potential anticancer profile. Eur J Med Chem156:461-478 (2018) [PubMed] Article
Channar, PA; Afzal, S; Ejaz, SA; Saeed, A; Larik, FA; Mahesar, PA; Lecka, J; Sévigny, J; Erben, MF; Iqbal, J Exploration of carboxy pyrazole derivatives: Synthesis, alkaline phosphatase, nucleotide pyrophosphatase/phosphodiesterase and nucleoside triphosphate diphosphohydrolase inhibition studies with potential anticancer profile. Eur J Med Chem156:461-478 (2018) [PubMed] Article