| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Glycylpeptide N-tetradecanoyltransferase 1/2 |
|---|
| Ligand | BDBM50034993 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_141306 (CHEMBL748920) |
|---|
| IC50 | 14100±n/a nM |
|---|
| Citation |  Devadas, B; Zupec, ME; Freeman, SK; Brown, DL; Nagarajan, S; Sikorski, JA; McWherter, CA; Getman, DP; Gordon, JI Design and syntheses of potent and selective dipeptide inhibitors of Candida albicans myristoyl-CoA:protein N-myristoyltransferase. J Med Chem38:1837-40 (1995) [PubMed] Article Devadas, B; Zupec, ME; Freeman, SK; Brown, DL; Nagarajan, S; Sikorski, JA; McWherter, CA; Getman, DP; Gordon, JI Design and syntheses of potent and selective dipeptide inhibitors of Candida albicans myristoyl-CoA:protein N-myristoyltransferase. J Med Chem38:1837-40 (1995) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Glycylpeptide N-tetradecanoyltransferase 1/2 |
|---|
| Name: | Glycylpeptide N-tetradecanoyltransferase 1/2 |
|---|
| Synonyms: | Peptide N-myristoyltransferase |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of ChEMBL is 141178 |
|---|
| Components: | This complex has 2 components. |
|---|
| Component 1 |
| Name: | Glycylpeptide N-tetradecanoyltransferase 2 |
|---|
| Synonyms: | Glycylpeptide N-tetradecanoyltransferase 2 | Myristoyl-CoA:protein N-myristoyltransferase 2 | NMT 2 | NMT2 | NMT2_HUMAN | Peptide N-myristoyltransferase 2 | Type II N-myristoyltransferase |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 56986.61 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_1435042 |
|---|
| Residue: | 498 |
|---|
| Sequence: | MAEDSESAASQQSLELDDQDTCGIDGDNEEETEHAKGSPGGYLGAKKKKKKQKRKKEKPN
SGGTKSDSASDSQEIKIQQPSKNPSVPMQKLQDIQRAMELLSACQGPARNIDEAAKHRYQ
FWDTQPVPKLDEVITSHGAIEPDKDNVRQEPYSLPQGFMWDTLDLSDAEVLKELYTLLNE
NYVEDDDNMFRFDYSPEFLLWALRPPGWLLQWHCGVRVSSNKKLVGFISAIPANIRIYDS
VKKMVEINFLCVHKKLRSKRVAPVLIREITRRVNLEGIFQAVYTAGVVLPKPIATCRYWH
RSLNPRKLVEVKFSHLSRNMTLQRTMKLYRLPDVTKTSGLRPMEPKDIKSVRELINTYLK
QFHLAPVMDEEEVAHWFLPREHIIDTFVVESPNGKLTDFLSFYTLPSTVMHHPAHKSLKA
AYSFYNIHTETPLLDLMSDALILAKSKGFDVFNALDLMENKTFLEKLKFGIGDGNLQYYL
YNWRCPGTDSEKVGLVLQ
|
|
|
|---|
| Component 2 |
| Name: | Glycylpeptide N-tetradecanoyltransferase 1 |
|---|
| Synonyms: | Myristoyl-CoA:protein N-myristoyltransferase (Nmt) | N-myristoyltransferase (NMT1) | NMT | NMT1 | NMT1_HUMAN | Peptide N-myristoyltransferase | Peptide N-myristoyltransferase 1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 56814.10 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P30419 |
|---|
| Residue: | 496 |
|---|
| Sequence: | MADESETAVKPPAPPLPQMMEGNGNGHEHCSDCENEEDNSYNRGGLSPANDTGAKKKKKK
QKKKKEKGSETDSAQDQPVKMNSLPAERIQEIQKAIELFSVGQGPAKTMEEASKRSYQFW
DTQPVPKLGEVVNTHGPVEPDKDNIRQEPYTLPQGFTWDALDLGDRGVLKELYTLLNENY
VEDDDNMFRFDYSPEFLLWALRPPGWLPQWHCGVRVVSSRKLVGFISAIPANIHIYDTEK
KMVEINFLCVHKKLRSKRVAPVLIREITRRVHLEGIFQAVYTAGVVLPKPVGTCRYWHRS
LNPRKLIEVKFSHLSRNMTMQRTMKLYRLPETPKTAGLRPMETKDIPVVHQLLTRYLKQF
HLTPVMSQEEVEHWFYPQENIIDTFVVENANGEVTDFLSFYTLPSTIMNHPTHKSLKAAY
SFYNVHTQTPLLDLMSDALVLAKMKGFDVFNALDLMENKTFLEKLKFGIGDGNLQYYLYN
WKCPSMGAEKVGLVLQ
|
|
|
|---|
| BDBM50034993 |
|---|
| n/a |
|---|
| Name | BDBM50034993 |
|---|
| Synonyms: | (S)-6-Amino-2-[(S)-3-hydroxy-2-(2-{4-[4-(2-methyl-imidazol-1-yl)-butyl]-phenyl}-acetylamino)-propionylamino]-hexanoic acid (2-cyclohexyl-ethyl)-amide | CHEMBL6269 | SC-58272 | [CYCLOHEXYLETHYL]-[[[[4-[2-METHYL-1-IMIDAZOLYL-BUTYL]PHENYL]ACETYL]-SERYL]-LYSINYL]-AMINE |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C33H52N6O4 |
|---|
| Mol. Mass. | 596.8038 |
|---|
| SMILES | Cc1nccn1CCCCc1ccc(CC(=O)N[C@@H](CO)C(=O)N[C@@H](CCCCN)C(=O)NCCC2CCCCC2)cc1 |
|---|
| Structure | 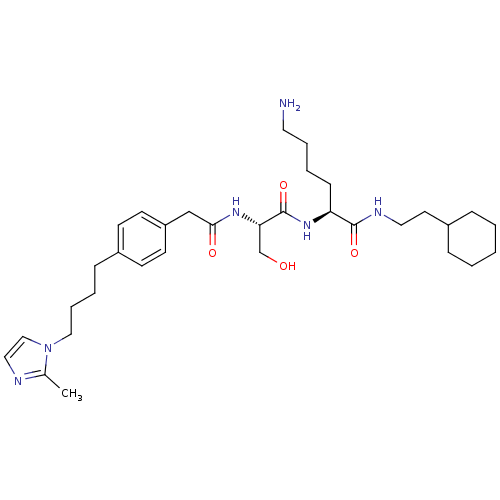
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Devadas, B; Zupec, ME; Freeman, SK; Brown, DL; Nagarajan, S; Sikorski, JA; McWherter, CA; Getman, DP; Gordon, JI Design and syntheses of potent and selective dipeptide inhibitors of Candida albicans myristoyl-CoA:protein N-myristoyltransferase. J Med Chem38:1837-40 (1995) [PubMed] Article
Devadas, B; Zupec, ME; Freeman, SK; Brown, DL; Nagarajan, S; Sikorski, JA; McWherter, CA; Getman, DP; Gordon, JI Design and syntheses of potent and selective dipeptide inhibitors of Candida albicans myristoyl-CoA:protein N-myristoyltransferase. J Med Chem38:1837-40 (1995) [PubMed] Article