

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Neprilysin | ||
| Ligand | BDBM50034860 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_144615 (CHEMBL750543) | ||
| IC50 | 515.0±n/a nM | ||
| Citation |  Ksander, GM; Ghai, RD; deJesus, R; Diefenbacher, CG; Yuan, A; Berry, C; Sakane, Y; Trapani, A Dicarboxylic acid dipeptide neutral endopeptidase inhibitors. J Med Chem38:1689-700 (1995) [PubMed] Ksander, GM; Ghai, RD; deJesus, R; Diefenbacher, CG; Yuan, A; Berry, C; Sakane, Y; Trapani, A Dicarboxylic acid dipeptide neutral endopeptidase inhibitors. J Med Chem38:1689-700 (1995) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Neprilysin | |||
| Name: | Neprilysin | ||
| Synonyms: | Mme | NEP_RAT | Neprilysin | Neutral Endopeptidase (NEP) | Neutral endopeptidase 24.11 | ||
| Type: | Protein | ||
| Mol. Mass.: | 85789.59 | ||
| Organism: | Rattus norvegicus (Rat) | ||
| Description: | P07861 | ||
| Residue: | 750 | ||
| Sequence: |
| ||
| BDBM50034860 | |||
| n/a | |||
| Name | BDBM50034860 | ||
| Synonyms: | 4-[4-Carboxy-2-(4-pyrrol-1-yl-benzyl)-5-(4-pyrrol-1-yl-phenyl)-pentanoylamino]-cyclohexanecarboxylic acid | CHEMBL45852 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C34H37N3O5 | ||
| Mol. Mass. | 567.6747 | ||
| SMILES | OC(=O)C(CC(Cc1ccc(cc1)-n1cccc1)C(=O)NC1CCC(CC1)C(O)=O)Cc1ccc(cc1)-n1cccc1 |(11.9,-3.3,;10.57,-2.53,;10.57,-.99,;9.22,-3.3,;7.89,-2.53,;6.56,-3.3,;6.56,-4.84,;5.79,-6.17,;6.54,-7.5,;5.76,-8.83,;4.21,-8.83,;3.45,-7.47,;4.23,-6.14,;3.43,-10.15,;4.04,-11.56,;2.89,-12.58,;1.57,-11.79,;1.9,-10.3,;5.23,-2.53,;5.23,-.99,;3.9,-3.3,;2.57,-2.53,;1.24,-3.3,;-.11,-2.53,;-.11,-.98,;1.24,-.22,;2.57,-.99,;-1.44,-.21,;-1.44,1.33,;-2.78,-.98,;9.22,-4.84,;10.31,-5.93,;9.92,-7.4,;11,-8.48,;12.47,-8.08,;12.86,-6.59,;11.78,-5.51,;13.56,-9.17,;13.33,-10.69,;14.71,-11.39,;15.8,-10.3,;15.1,-8.92,)| | ||
| Structure | 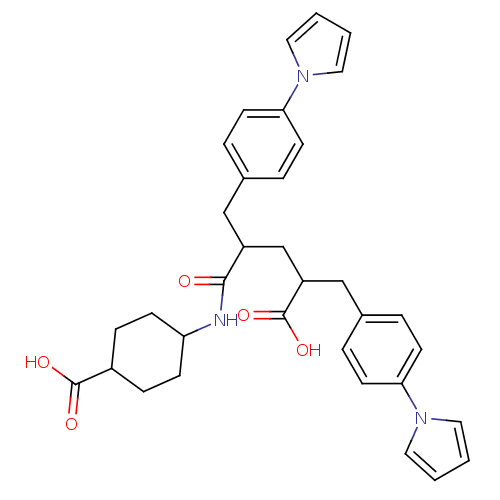
| ||