| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Endolysin |
|---|
| Ligand | BDBM26187 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_941016 (CHEMBL2330742) |
|---|
| Kd | 103000±n/a nM |
|---|
| Citation |  Merski, M; Shoichet, BK The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition. J Med Chem56:2874-84 (2013) [PubMed] Article Merski, M; Shoichet, BK The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition. J Med Chem56:2874-84 (2013) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Endolysin |
|---|
| Name: | Endolysin |
|---|
| Synonyms: | 3.2.1.17 | E | ENLYS_BPT4 | Endolysin | Lysis protein | Lysozyme | Muramidase |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 18699.91 |
|---|
| Organism: | Enterobacteria phage T4 |
|---|
| Description: | ChEMBL_100178 |
|---|
| Residue: | 164 |
|---|
| Sequence: | MNIFEMLRIDERLRLKIYKDTEGYYTIGIGHLLTKSPSLNAAKSELDKAIGRNCNGVITK
DEAEKLFNQDVDAAVRGILRNAKLKPVYDSLDAVRRCALINMVFQMGETGVAGFTNSLRM
LQQKRWDEAAVNLAKSIWYNQTPNRAKRVITTFRTGTWDAYKNL
|
|
|
|---|
| BDBM26187 |
|---|
| n/a |
|---|
| Name | BDBM26187 |
|---|
| Synonyms: | α-CA inhibitor, 11 | CHEMBL14060 | US9688816, 2 | phenol |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C6H6O |
|---|
| Mol. Mass. | 94.1112 |
|---|
| SMILES | Oc1ccccc1 |
|---|
| Structure | 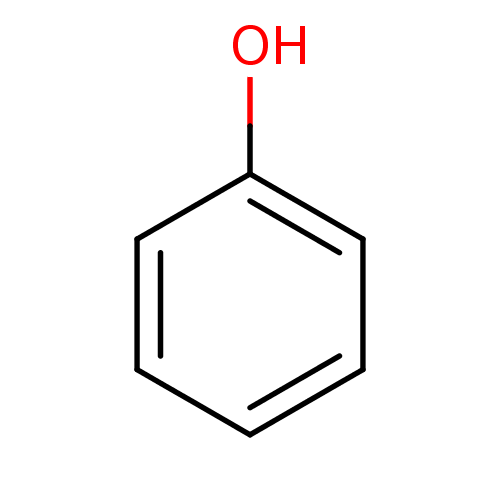
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Merski, M; Shoichet, BK The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition. J Med Chem56:2874-84 (2013) [PubMed] Article
Merski, M; Shoichet, BK The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition. J Med Chem56:2874-84 (2013) [PubMed] Article