| Reaction Details |
|---|
 | Report a problem with these data |
| Target | DNA ligase 1 |
|---|
| Ligand | BDBM25525 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1837505 (CHEMBL4337638) |
|---|
| IC50 | 382000±n/a nM |
|---|
| Citation |  Saquib, M; Ansari, MI; Johnson, CR; Khatoon, S; Kamil Hussain, M; Coop, A Recent advances in the targeting of human DNA ligase I as a potential new strategy for cancer treatment. Eur J Med Chem182:0 (2019) [PubMed] Article Saquib, M; Ansari, MI; Johnson, CR; Khatoon, S; Kamil Hussain, M; Coop, A Recent advances in the targeting of human DNA ligase I as a potential new strategy for cancer treatment. Eur J Med Chem182:0 (2019) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| DNA ligase 1 |
|---|
| Name: | DNA ligase 1 |
|---|
| Synonyms: | ATP-dependent DNA ligase | DNA Ligase I | DNLI1_HUMAN | Fatty Acid Amide Hydrolase | LIG1 | Polydeoxyribonucleotide synthase [ATP] |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 101721.76 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 919 |
|---|
| Sequence: | MQRSIMSFFHPKKEGKAKKPEKEASNSSRETEPPPKAALKEWNGVVSESDSPVKRPGRKA
ARVLGSEGEEEDEALSPAKGQKPALDCSQVSPPRPATSPENNASLSDTSPMDSSPSGIPK
RRTARKQLPKRTIQEVLEEQSEDEDREAKRKKEEEEEETPKESLTEAEVATEKEGEDGDQ
PTTPPKPLKTSKAETPTESVSEPEVATKQELQEEEEQTKPPRRAPKTLSSFFTPRKPAVK
KEVKEEEPGAPGKEGAAEGPLDPSGYNPAKNNYHPVEDACWKPGQKVPYLAVARTFEKIE
EVSARLRMVETLSNLLRSVVALSPPDLLPVLYLSLNHLGPPQQGLELGVGDGVLLKAVAQ
ATGRQLESVRAEAAEKGDVGLVAENSRSTQRLMLPPPPLTASGVFSKFRDIARLTGSAST
AKKIDIIKGLFVACRHSEARFIARSLSGRLRLGLAEQSVLAALSQAVSLTPPGQEFPPAM
VDAGKGKTAEARKTWLEEQGMILKQTFCEVPDLDRIIPVLLEHGLERLPEHCKLSPGIPL
KPMLAHPTRGISEVLKRFEEAAFTCEYKYDGQRAQIHALEGGEVKIFSRNQEDNTGKYPD
IISRIPKIKLPSVTSFILDTEAVAWDREKKQIQPFQVLTTRKRKEVDASEIQVQVCLYAF
DLIYLNGESLVREPLSRRRQLLRENFVETEGEFVFATSLDTKDIEQIAEFLEQSVKDSCE
GLMVKTLDVDATYEIAKRSHNWLKLKKDYLDGVGDTLDLVVIGAYLGRGKRAGRYGGFLL
ASYDEDSEELQAICKLGTGFSDEELEEHHQSLKALVLPSPRPYVRIDGAVIPDHWLDPSA
VWEVKCADLSLSPIYPAARGLVDSDKGISLRFPRFIRVREDKQPEQATTSAQVACLYRKQ
SQIQNQQGEDSGSDPEDTY
|
|
|
|---|
| BDBM25525 |
|---|
| n/a |
|---|
| Name | BDBM25525 |
|---|
| Synonyms: | 24-methyl-5,7,18,20-tetraoxa-24-azahexacyclo[11.11.0.0^{2,10}.0^{4,8}.0^{14,22}.0^{17,21}]tetracosa-1(13),2,4(8),9,11,14(22),15,17(21),23-nonaen-24-ium | CHEMBL490129 | Pseudochelerythrine | Veadent | cid_5154 | sanguinarine | sangvinarin |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H14NO4 |
|---|
| Mol. Mass. | 332.3289 |
|---|
| SMILES | C[n+]1cc2c3OCOc3ccc2c2ccc3cc4OCOc4cc3c12 |
|---|
| Structure | 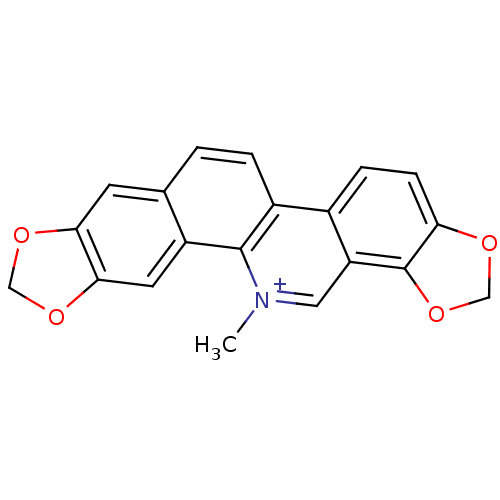
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Saquib, M; Ansari, MI; Johnson, CR; Khatoon, S; Kamil Hussain, M; Coop, A Recent advances in the targeting of human DNA ligase I as a potential new strategy for cancer treatment. Eur J Med Chem182:0 (2019) [PubMed] Article
Saquib, M; Ansari, MI; Johnson, CR; Khatoon, S; Kamil Hussain, M; Coop, A Recent advances in the targeting of human DNA ligase I as a potential new strategy for cancer treatment. Eur J Med Chem182:0 (2019) [PubMed] Article