| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Serine protease 1 |
|---|
| Ligand | BDBM50066641 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_212528 (CHEMBL817592) |
|---|
| Ki | 310±n/a nM |
|---|
| Citation |  Phillips, GB; Buckman, BO; Davey, DD; Eagen, KA; Guilford, WJ; Hinchman, J; Ho, E; Koovakkat, S; Liang, A; Light, DR; Mohan, R; Ng, HP; Post, JM; Shaw, KJ; Smith, D; Subramanyam, B; Sullivan, ME; Trinh, L; Vergona, R; Walters, J; White, K; Whitlow, M; Wu, S; Xu, W; Morrissey, MM Discovery of N-[2-[5-[Amino(imino)methyl]-2-hydroxyphenoxy]-3, 5-difluoro-6-[3-(4, 5-dihydro-1-methyl-1H-imidazol-2-yl)phenoxy]pyridin-4-yl]-N-methylgl y cine (ZK-807834): a potent, selective, and orally active inhibitor of the blood coagulation enzyme factor Xa. J Med Chem41:3557-62 (1998) [PubMed] Article Phillips, GB; Buckman, BO; Davey, DD; Eagen, KA; Guilford, WJ; Hinchman, J; Ho, E; Koovakkat, S; Liang, A; Light, DR; Mohan, R; Ng, HP; Post, JM; Shaw, KJ; Smith, D; Subramanyam, B; Sullivan, ME; Trinh, L; Vergona, R; Walters, J; White, K; Whitlow, M; Wu, S; Xu, W; Morrissey, MM Discovery of N-[2-[5-[Amino(imino)methyl]-2-hydroxyphenoxy]-3, 5-difluoro-6-[3-(4, 5-dihydro-1-methyl-1H-imidazol-2-yl)phenoxy]pyridin-4-yl]-N-methylgl y cine (ZK-807834): a potent, selective, and orally active inhibitor of the blood coagulation enzyme factor Xa. J Med Chem41:3557-62 (1998) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Serine protease 1 |
|---|
| Name: | Serine protease 1 |
|---|
| Synonyms: | Beta-Trypsin | Cationic trypsin | PRSS1 | TRP1 | TRY1 | TRY1_BOVIN | TRYP1 | Trypsin | Trypsin I |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 25790.52 |
|---|
| Organism: | Bos taurus (bovine) |
|---|
| Description: | P00760 |
|---|
| Residue: | 246 |
|---|
| Sequence: | MKTFIFLALLGAAVAFPVDDDDKIVGGYTCGANTVPYQVSLNSGYHFCGGSLINSQWVVS
AAHCYKSGIQVRLGEDNINVVEGNEQFISASKSIVHPSYNSNTLNNDIMLIKLKSAASLN
SRVASISLPTSCASAGTQCLISGWGNTKSSGTSYPDVLKCLKAPILSDSSCKSAYPGQIT
SNMFCAGYLEGGKDSCQGDSGGPVVCSGKLQGIVSWGSGCAQKNKPGVYTKVCNYVSWIK
QTIASN
|
|
|
|---|
| BDBM50066641 |
|---|
| n/a |
|---|
| Name | BDBM50066641 |
|---|
| Synonyms: | (2S,4R)-4-{2-(5-Carbamimidoyl-2-hydroxy-phenoxy)-3,5-difluoro-6-[3-(1-methyl-4,5-dihydro-1H-imidazol-2-yl)-phenoxy]-pyridin-4-yloxy}-pyrrolidine-2-carboxylic acid | CHEMBL332189 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H26F2N6O6 |
|---|
| Mol. Mass. | 568.5287 |
|---|
| SMILES | CN1CCN=C1c1cccc(Oc2nc(Oc3cc(ccc3O)C(N)=N)c(F)c(O[C@H]3CN[C@@H](C3)C(O)=O)c2F)c1 |c:4| |
|---|
| Structure | 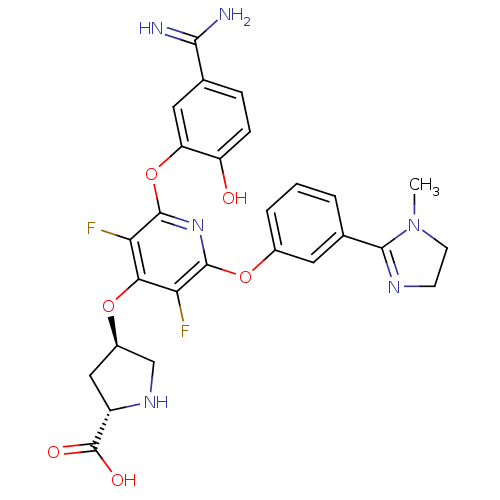
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Phillips, GB; Buckman, BO; Davey, DD; Eagen, KA; Guilford, WJ; Hinchman, J; Ho, E; Koovakkat, S; Liang, A; Light, DR; Mohan, R; Ng, HP; Post, JM; Shaw, KJ; Smith, D; Subramanyam, B; Sullivan, ME; Trinh, L; Vergona, R; Walters, J; White, K; Whitlow, M; Wu, S; Xu, W; Morrissey, MM Discovery of N-[2-[5-[Amino(imino)methyl]-2-hydroxyphenoxy]-3, 5-difluoro-6-[3-(4, 5-dihydro-1-methyl-1H-imidazol-2-yl)phenoxy]pyridin-4-yl]-N-methylgl y cine (ZK-807834): a potent, selective, and orally active inhibitor of the blood coagulation enzyme factor Xa. J Med Chem41:3557-62 (1998) [PubMed] Article
Phillips, GB; Buckman, BO; Davey, DD; Eagen, KA; Guilford, WJ; Hinchman, J; Ho, E; Koovakkat, S; Liang, A; Light, DR; Mohan, R; Ng, HP; Post, JM; Shaw, KJ; Smith, D; Subramanyam, B; Sullivan, ME; Trinh, L; Vergona, R; Walters, J; White, K; Whitlow, M; Wu, S; Xu, W; Morrissey, MM Discovery of N-[2-[5-[Amino(imino)methyl]-2-hydroxyphenoxy]-3, 5-difluoro-6-[3-(4, 5-dihydro-1-methyl-1H-imidazol-2-yl)phenoxy]pyridin-4-yl]-N-methylgl y cine (ZK-807834): a potent, selective, and orally active inhibitor of the blood coagulation enzyme factor Xa. J Med Chem41:3557-62 (1998) [PubMed] Article